Microbial contamination, including the carryover of infectious microbes, is a global public health concern. An alternative technique that serves as a powerful, rapid, and highly sensitive method for the specific and non-specific detection, monitoring, enumeration, and characterization of microorganisms is flow cytometry (FCM).
- flow cytometry
- fluorescence in situ hybridization
- microbial contamination
1. Introduction
Microbial resistance against extrinsic factors is related to their fast adaptability and the formation of microbial biofilms, which can protect spoilage microorganisms and bacterial pathogens from chemical and physical actions [2,3,5,6][1][2][3][4]. Frequently detected pathogens include Salmonella spp., Listeria monocytogenes, Escherichia coli, Shigella spp., Vibrio spp., Campylobacter jejuni, and Yersinia spp., whereas frequent spoilage microorganisms are, for instance, Acinetobacter spp., Pseudomonas spp. or Botrytis spp. [7,8,9,10][5][6][7][8].
Effective qualitative and quantitative monitoring and detection tools are required to minimize the contamination risk. The gold standard among detection tools is still the conventional plating method, with its high sensitivity and selectivity [12,13][9][10]. However, plating is time-consuming, labor-intensive, and detects only viable and cultivable microbes [7,14][5][11]. Complementarily, there are several rapid and culture-independent approaches that overcome these limitations of plating. Among the most widely-known detection methods are molecular methods such as polymerase chain reaction (PCR) or enzyme-linked immunosorbent assay (ELISA) methods [7,13,15,16][5][10][12][13]. Some molecular methods are vulnerable to interference from inhibitory compounds (i.e., the lipid content) or can affect complex matrices such as food. For highly sensitive methods such as PCR, contamination can easily lead to false results. In addition, PCR may be unable to distinguish between viable and non-viable cells [13,17][10][14].
Flow cytometry (FCM) allows a culture-independent quantitative count of microbial cells. In addition, flow cytometry provides information on the physiological and structural characteristics of microbial cells and their viability and can therefore be used as an additional characterization tool. Rapid and reliable detection, quantification and characterization of foodborne pathogens are of great interest to the food industry in order to minimize foodborne diseases [19][15]. The rapid techniques used to detect foodborne pathogens can be categorized into immunological, biosensor, and nucleic acid-based methods [20][16]. Fluorescence in situ hybridization (FISH) is a nucleic acid-based method and is mainly applied in the medical and diagnostic field [19][15].
2. Non-Specific State-of-the-Art Flow Cytometric Applications for Detection and Monitoring
2.1. FCM Principle and Detection Mechanisms
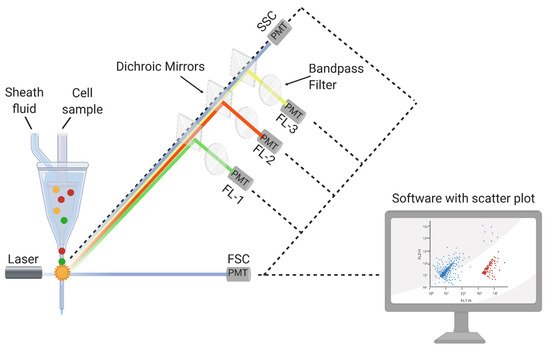
3.2. Food-Related FCM Applications
2.2. Food-Related FCM Applications
43. Specific State-of-the-Art Flow-FISH Methods and Applications for Monitoring and Detection
4.1. Principle of FISH
3.1. Principle of FISH
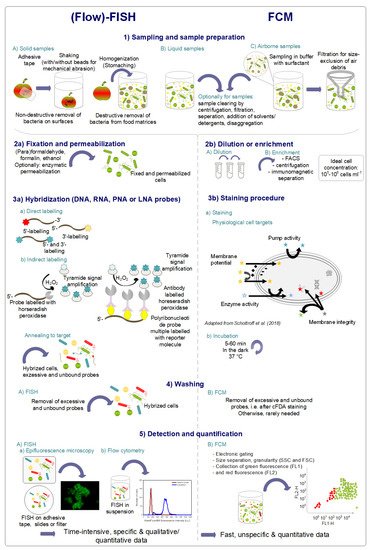
4.2. Flow-FISH in Food Microbiology
3.2. Flow-FISH in Food Microbiology
References
- Havelaar, A.H.; Brul, S.; De Jong, A.; De Jonge, R.; Zwietering, M.H.; Ter Kuile, B.H. Future challenges to microbial food safety. Int. J. Food Microbiol. 2010, 139, S79–S94.
- Juzwa, W.; Duber, A.; Myszka, K.; Bialas, W.; Czaczyk, K. Identification of microbes from the surfaces of food-processing lines based on the flow cytometric evaluation of cellular metabolic activity combined with cell sorting. Biofouling 2016, 32, 841–851.
- Wang, R. Biofilms and meat safety: A mini-review. J. Food Prot. 2019, 82, 120–127.
- Weber, M.; Liedtke, J.; Plattes, S.; Lipski, A. Bacterial community composition of biofilms in milking machines of two dairy farms assessed by a combination of culture-dependent and –independent methods. PLoS ONE 2019, 14, e0222238.
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73.
- Marušić, A. Food safety and security: What were favourite topics for research in the last decade? J. Glob. Health 2011, 1, 72–78.
- Pepe, T.; De Dominicis, R.; Esposito, G.; Ventrone, I.; Fratamico, P.M.; Cortesi, M.L. Detection of Campylobacter from poultry carcass skin samples at slaughter in Southern Italy. J. Food Prot. 2009, 72, 1718–1721.
- Rohde, A.; Hammerl, J.A.; Al Dahouk, S. Detection of foodborne bacterial zoonoses by fluorescence in situ hybridization. Food Control 2016, 69, 297–305.
- Ge, B.; Meng, J. Advanced technologies for pathogen and toxin detection in foods: Current applications and future directions. JALA J. Assoc. Lab. Autom. 2009, 14, 235–241.
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, 396–411.
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254.
- Ricke, S.C.; Feye, K.M.; Chaney, W.E.; Shi, Z.; Pavlidis, H.; Yang, Y. Developments in rapid detection methods for the detection of foodborne campylobacterin the United States. Front. Microbiol. 2019, 10, 3280.
- Wei, Q.Y.; Wang, X.M.; Sun, D.W.; Pu, H.B. Rapid detection and control of psychrotrophic microorganisms in cold storage foods: A review. Trends Food Sci. Technol. 2019, 86, 453–464.
- Safford, H.R.; Bischel, H.N. Flow cytometry applications in water treatment, distribution, and reuse: A review. Water Res. 2019, 151, 110–133.
- Dias, G.; Rathnayaka, U. Fluorescence in situ hybridization (FISH) in food pathogen detection. Int. J. Mol. Biol. 2018, 3, 143–149.
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 770.
- Comas-Riu, J.; Rius, N. Flow cytometry applications in the food industry. J. Ind. Microbiol. Biotechnol. 2009, 36, 999–1011.
- Paparella, A.; Serio, A.; Chaves, C. Flow cytometry applications in food safety studies. Flow Cytom.-Recent Perspect. 2012, 69–102.
- Veal, D.A.; Deere, D.; Ferrari, B.; Piper, J.; Attfield, P.V. Fluorescence staining and flow cytometry for monitoring microbial cells. J. Immunol. Methods 2000, 243, 191–210.
- Wu, L.N.; Wang, S.; Song, Y.Y.; Wang, X.; Yan, X.M. Applications and challenges for single-bacteria analysis by flow cytometry. Sci. China-Chem. 2016, 59, 30–39.
- Wilkinson, M.G. Flow cytometry as a potential method of measuring bacterial viability in probiotic products: A review. Trends Food Sci. Technol. 2018, 78, 1–10.
- Tracy, B.P.; Gaida, S.M.; Papoutsakis, E.T. Flow cytometry for bacteria: Enabling metabolic engineering, synthetic biology and the elucidation of complex phenotypes. Curr. Opin. Biotechnol. 2010, 21, 85–99.
- Barros, C.P.; Pires, R.P.S.; Guimarães, J.T.; Abud, Y.K.D.; Almada, C.N.; Pimentel, T.C.; Sant’Anna, C.; De-Melo, L.D.B.; Duarte, M.C.K.H.; Silva, M.C.; et al. Ohmic heating as a method of obtaining paraprobiotics: Impacts on cell structure and viability by flow cytometry. Food Res. Int. 2021, 140, 110061.
- Braschi, G.; Patrignani, F.; Siroli, L.; Lanciotti, R.; Schlueter, O.; Froehling, A. Flow Cytometric Assessment of the Morphological and Physiological Changes of Listeria monocytogenes and Escherichia coli in Response to Natural Antimicrobial Exposure. Front. Microbiol. 2018, 9, 2783.
- Fröhling, A.; Baier, M.; Ehlbeck, J.; Knorr, D.; Schlüter, O. Atmospheric pressure plasma treatment of Listeria innocua and Escherichia coli at polysaccharide surfaces: Inactivation kinetics and flow cytometric characterization. Innov. Food Sci. Emerg. Technol. 2012, 13, 142–150.
- Fröhling, A.; Wienke, M.; Rose-Meierhofer, S.; Schlüter, O. Improved method for mastitis detection and evaluation of disinfectant efficiency during milking process. Food Bioprocess Technol. 2010, 3, 892–900.
- Jaeger, H.; Schulz, A.; Karapetkov, N.; Knorr, D. Protective effect of milk constituents and sublethal injuries limiting process effectiveness during PEF inactivation of Lb. rhamnosus. Int. J. Food Microbiol. 2009, 134, 154–161.
- Schenk, M.; Raffellini, S.; Guerrero, S.; Blanco, G.A.; Alzamora, S.M. Inactivation of Escherichia coli, Listeria innocua and Saccharomyces cerevisiae by UV-C light: Study of cell injury by flow cytometry. LWT-Food Sci. Technol. 2011, 44, 191–198.
- Tamburini, S.; Ballarini, A.; Ferrentino, G.; Moro, A.; Foladori, P.; Spilimbergo, S.; Jousson, O. Comparison of quantitative PCR and flow cytometry as cellular viability methods to study bacterial membrane permeabilization following supercritical CO2 treatment. Microbiology 2013, 159, 1056–1066.
- Teixeira, P.; Fernandes, B.; Silva, A.M.; Dias, N.; Azeredo, J. Evaluation by flow cytometry of Escherichia coli viability in lettuce after disinfection. Antibiotics 2020, 9, 14.
- Zand, E.; Schottroff, F.; Steinacker, E.; Mae-Gano, J.; Schoenher, C.; Wimberger, T.; Wassermann, K.J.; Jaeger, H. Advantages and limitations of various treatment chamber designs for reversible and irreversible electroporation in life sciences. Bioelectrochemistry 2021, 141, 107841.
- Fröhling, A.; Schlüter, O. Flow cytometric evaluation of physico-chemical impact on Gram-positive and Gram-negative bacteria. Front. Microbiol. 2015, 6, 939.
- Khan, M.M.T.; Pyle, B.H.; Camper, A.K. Specific and Rapid Enumeration of Viable but Nonculturable and Viable-Culturable Gram-Negative Bacteria by Using Flow Cytometry. Appl. Environ. Microbiol. 2010, 76, 5088–5096.
- Mao, C.; Xue, C.; Wang, X.; He, S.; Wu, L.; Yan, X. Rapid quantification of pathogenic Salmonella Typhimurium and total bacteria in eggs by nano-flow cytometry. Talanta 2020, 217, 121020.
- Yu, M.X.; Wu, L.N.; Huang, T.X.; Wang, S.; Yan, X.M. Rapid detection and enumeration of total bacteria in drinking water and tea beverages using a laboratory-built high-sensitivity flow cytometer. Anal. Methods 2015, 7, 3072–3079.
- Coronel-Leon, J.; Lopez, A.; Espuny, M.J.; Beltran, M.T.; Molinos-Gomez, A.; Rocabayera, X.; Manresa, A. Assessment of antimicrobial activity of N-alpha-lauroyl arginate ethylester (LAE (R)) against Yersinia enterocolitica and Lactobacillus plantarum by flow cytometry and transmission electron microscopy. Food Control 2016, 63, 1–10.
- Carrillo, M.G.; Ferrario, M.; Guerrero, S. Effectiveness of UV-C light assisted by mild heat on Saccharomyces cerevisiae KE 162 inactivation in carrot-orange juice blend studied by flow cytometry and transmission electron microscopy. Food Microbiol. 2018, 73, 1–10.
- Barer, M.R.; Gribbon, L.T.; Harwood, C.R.; Nwoguh, C.E. The viable but non-culturable hypothesis and medical bacteriology. Rev. Med Microbiol. 1993, 4, 183–191.
- DeLong, E.F.; Wickham, G.S.; Pace, N.R. Phylogenetic stains: Ribosomal RNA-based probes for the identification of single cells. Science 1989, 243, 1360–1363.
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S ribosomal-RNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925.
- Oliveira, R.; Almeida, C.; Azevedo, N.F. Detection of microorganisms by fluorescence in situ hybridization using peptide nucleic acid. In Peptide Nucleic Acids: Methods and Protocols; Nielsen, P.E., Ed.; Springer: New York, NY, USA, 2020; pp. 217–230.
- Wagner, M.; Haider, S. New trends in fluorescence in situ hybridization for identification and functional analyses of microbes. Curr. Opin. Biotechnol. 2012, 23, 96–102.
- Amann, R.; Fuchs, B.M.; Behrens, S. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 2001, 12, 231–236.
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348.
- Rohde, A.; Hammerl, J.A.; Appel, B.; Dieckmann, R.; Al Dahouk, S. FISHing for bacteria in food—A promising tool for the reliable detection of pathogenic bacteria? Food Microbiol. 2015, 46, 395–407.
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol. Rev. 1995, 59, 143–169.
- Bokulich, N.A.; Mills, D.A. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 2012, 45, 377–389.
- Amann, R.I.; Krumholz, L.; Stahl, D.A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 1990, 172, 762–770.
- Gunasekera, T.S.; Veal, D.A.; Attfield, P.V. Potential for broad applications of flow cytometry and fluorescence techniques in microbiological and somatic cell analyses of milk. Int. J. Food Microbiol. 2003, 85, 269–279.
- Zwirglmaier, K. Fluorescence in situ hybridisation (FISH)—The next generation. FEMS Microbiol. Lett. 2005, 246, 151–158.
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Correction: Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS ONE 2013, 8, e14786.
- Šilhová, L.; Moťková, P.; Šilha, D.; Vytřasová, J. FISH detection of Campylobacter and Arcobacter adhered to stainless steel coupons. J. Microbiol. Biotechnol. Food Sci. 2015, 4, 347–351.
- Bragança, S.M.; Azevedo, N.F.; Simões, L.C.; Keevil, C.W.; Vieira, M.J. Use of fluorescent in situ hybridisation for the visualisation of Helicobacter pylori in real drinking water biofilms. Water Sci. Technol. 2007, 55, 387–393.
