Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Vivi Li and Version 1 by Mickael Bourge.
Cotoneaster integerrimus represents a multiploid and facultative apomictic system of widely distributed mountain populations.
- apomixis
- Balkans
- clonality
- Cotoneaster integerrimus
- cytotype
- genome size
- polyploidy
1. Introduction
Genome size, as a fundamental biological characteristic of evolutionary significance, has implications on overall biodiversity [1]. The causes of genome size variation in plants are varied, but the most striking is polyploidization. The evolutionary significance of polyploidy has tremendous and far-reaching implications for plant diversity [2]. It is assumed that almost 50% of plant groups have been affected by at least a single polyploidization event throughout their evolutionary history [3]. Polyploidy recomposes genome structure, alters gene expression, induces phenotypic and physiological changes, and provide adaptive potential to the polyploid plant [4,5,6][4][5][6]. Allopolyploidy is considered to be more frequent than autopolyploidy [7], but both processes profoundly shape plant diversity, either independently or in concert ([8], and references therein).
One of the consequences of being a polyploid is breakdown of self-incompatibility, allowing a shift towards asexual reproductive mode [9,10,11][9][10][11]. Apomixis (asexual seed formation) represents an effective life strategy by which apomicts preserve and maintain hybrid and heterozygous genetic lineages, cytotypes with unbalanced chromosomes, enabling their long-term persistence and dispersal [12]. Apomixis is highly correlated with polyploidy and hybridity. However, it is still unclear as to which of these factors precedes and what exact genetic and developmental mechanisms regulate the emergence of apomixis [13].
Gametophytic apomixis, the most common type of asexual reproduction, represents the formation and development of unreduced megagametophyte (parthenogenesis) coupled with the fertilization of unreduced central cell (pseudogamy) [14,15][14][15]. As a consequence of bypassing meiosis and lack of genome recombination, apomicts have a genetic structure identical to the maternal plant, thus creating clonal populations. Apomictic polyploids show a greater colonization ability by occupying more extreme ecological niches and persisting in larger distribution areas (geographic parthenogenesis, [14]) than their diploid sexual relatives [16,17,18][16][17][18]. Although genetically uniform, apomicts acquire genetic variation over time via accumulation of spontaneous mutations and via residual sexuality fostering diversity within clonal populations [19,20,21][19][20][21]. One of the mechanisms enhancing genetic diversity of apomictic populations comes from crosses between asexual and related sexual lineages, resulting in offspring that have a predominant apomictic mode of reproduction [17,22,23][17][22][23]. Given the long-life cycle of woody species, apomictic reproduction and independence of mates is extremely important in the early stages of the establishment of new populations and colonization of new environments.
Genus Cotoneaster Medik. (Rosaceae, Spiroaeoideae, Pyreae, [24]) includes shrubs and some small trees that are distributed in Europe, North Africa, and temperate areas of Asia (except Japan) [25]. The species number varies greatly among authors and ranges from 50–70 [26], to ≈90 [27], to ≈400 [25]. Evolutionary history and diversification of Cotoneaster have been affected by homoploid and heteroploid hybridization, as well as autopolyploidy and apomixis [25,28,29,30,31,32,33,34,35][25][28][29][30][31][32][33][34][35]. Both classical chromosome countings and cytometric analyses confirmed a series of ploidy levels in the genus ranging from diploids to hexaploids, with a prevalence of tetraploids.
Apomixis has been proven to be the most frequent reproductive mode in Cotoneaster that was primarily inferred from embryological data [36[36][37][38],37,38], breeding experiments [39], and later by molecular analyses [31,33][31][33]. Recently, apomixis has been confirmed using flow cytometry and different reproductive pathways of seed formation in tetraploids of C. integerrimus Medik. [29,30][29][30].
Key information on embryo sac development of several tri- and tetraploid Cotoneaster species (C. rosea Edgew., C. nitens Rehd. & Wils., C. bullata Bois, C. obscura Rehd. & Wils., C. acutifolia var. villosula Rehd. & Wils., C. racemiflora var. soongorica Schneid.) was provided in 1962 [36], and for C. melanocarpus Fisch. ex Blytt much later [37,38][37][38]. These species showed a strong tendency towards apomictic reproduction, namely, apospory and rarely diplospory, and the early development of their embryo sacs had general similarities with the other apomictic rosaceous genera. The most frequent development of unreduced embryo sac implied degeneration of the primary megaspore mother cell in the earliest developmental stages of the ovule and its replacement with unreduced embryo sac originating from nucellar cells (apospory). In several cases, the unreduced embryo sac developed from archesporial cells following the degeneration of the primary megaspore mother cell (diplospory). The secondary mother megaspore cell also occurred and was developed from unreduced embryo sac or via meiosis [36]. The reduced embryo sacs were rarely observed in the studied species [36,37,38][36][37][38]. The authors did not provide data on the polar nuclei. Conclusion of those findings is that most polyploid species of Cotoneaster are apomictic [25,32][25][32]. Sexual reproduction is less represented in Cotoneaster and follows a common pattern of development of Polygonum type of embryo sac: seeds of diploid mothers have diploid embryo and triploid endosperm; seeds of tetraploid mothers have tetraploid embryo and hexaploid endosperm [29,30][29][30].
In recent studies, only a tetraploid cytotype in C. integerrimus populations from Central Europe was found [30,40][30][40]. The same authors [30] proved that facultative apomixis was the main reproductive mode, followed by autonomous apomixis and haploid parthenogenesis, while the proportion of sexuality was 10% in the studied sample. The authors showed that different reproductive pathways involved the interaction of reduced and unreduced gametes documented in both sexuals and asexuals.
2. Genome Size and Ploidy Level Variation
The flow cytometry (FCM) of 208 Cotoneaster integerrimus individuals from Bosnia and Herzegovina resulted in three distinct groups whose holoploid genome sizes (2C) corresponded to three different ploidy levels, namely, di-, tri- and tetraploid cytotypes (Table 1, Figure 1A). The two singular values corresponded to pentaploid (2C = 2.99 pg, 1Cx = 0.598 pg) and hexaploid cytotype (2C = 3.35 pg, 1Cx = 0.558 pg); thus, both values were excluded from further analyses. The 2C genome size mean values were 1.28 pg in diploids, 1.856 pg in triploids, and 2.481 pg in tetraploids. The significantly (F2, 207 = 10.98, p > 0.001) highest monoploid (1Cx) genome size value was recorded in diploid, and the lowest in triploid cytotype (Table 1).
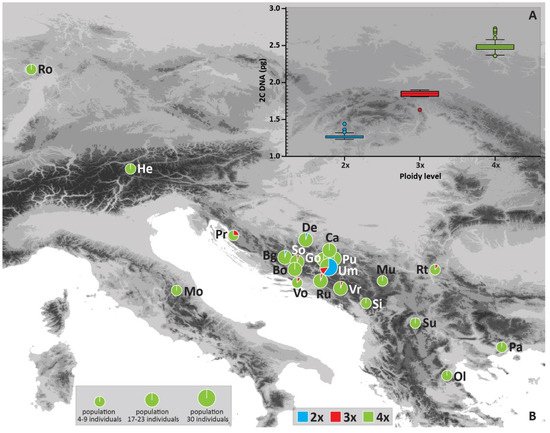

Figure 1. Absolute nuclear genome size (2C) of diploid, triploid, and tetraploid cytotypes (A) (boxes define 95th percentile interval; horizontal lines indicate mean value; whiskers indicate standard deviation: outliers are shown as circles), and geographical distribution of Cotoneaster integerrimus cytotypes and their frequency (%) per site/population (B). Site abbreviations (Site IDs) correspond to Table 2.
Table 1. Absolute genome size values and ploidy levels of Cotoneaster integerrimus cytotypes.
| DNA Ploidy | N | 2C DNA | CV % | Cytometric Results | 1Cx DNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD (pg) | Min–Max (pg) | Mean ± SD (pg) * | Mean (Mbp **) | |||||||||
| Hypothesized Pathways of Seed Formation | ||||||||||||
| 2x | 20 | 1.280 ± 0.046 | 1.23–1.36 | 2.47 | 0.636 ± 0.015 | a | 622.00 | |||||
| Maternal ploidy | a | Embryopg ± SD | Endosperm pg ± SD | Embryo ploidy | Endosperm ploidy | Number of seeds | Egg ploidy | Polar nuclei ploidy/number | ||||
| 3x | 7 | 1.856 ± 0.091 | 1.63–1.90 | 5.04 | 0.608 ± 0.030 | b | 594.62 | |||||
| 4x | 181 | 2.481 ± 0.060 | 2.36–2.73 | 2.45 | 0.621 ± 0.015 | b | 607.73 | |||||
| TOTAL | 208 | |||||||||||
* Mean ± SD (pg) followed by the same letter in superscript was not significantly different according to the Tukey’s HSD test; ** 1pg = 978 Mbp [41].
Table 2. Geographic origin of Cotoneaster mother individuals covering three cytotypes and corresponding sample sizes (N) for genome size/ploidy level, reproductive mode, and microsatellite analysis.
| Origin of Sample | Number of Samples ( | N | ) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site No. | Site ID | Locality | East | North | Altitude (m) | Diploids | Triploids | Tetraploids | Total | N | per Site | ||||||
| Sperm ploidy | Number of sperms fecundating the endosperm | Endosperm maternal: paternal genome ratio | Endosperm/embryo fluorescence (mean ratio) | Seed origin | 1 | Go | |||||||||||
| 2x | 1.28 ± 0.03 | Gornja Grkarica, Mt. Igman, Bosnia and Herzegovina (B&H) | 18.28833 | ||||||||||||||
| 1 | |||||||||||||||||
| 3x | 3x/2 | 3x | 1 | 2m:1p | 2.12 | Pseudogamous Apomixis | |||||||||||
| 4x | 2.54 ± 0.07 | 3.84 ± 0.11 | 4x | 6x | 92 | 2x | 2x/2 | 2x | 1 | 2m:1p | 1.53 | Sexual | |||||
| 4x | 3.80 ± 0.10 | 6.43 ± 0.21 | 6x | 10x | 22 | c | 4x | 4x/2 | 2x | 2 | 4m:1p | 2.5 | Sexual | ||||
| 4x | 1.90 ± 0.01 | 3.19 ± 0.05 | 3x | 5x | 2 | d | 2x | 2x/1 | 1x | 1 | 4m:1p | 1.7 | Sexual | ||||
| 4x | 2.56 ± 0.1 | 6.38 ± 0.15 | 4x | 10x | 148 | 4x | 4x/2 | 1x or 2x | 2 or 1 | 4m:1p | 2.5 | Pseudogamous Apomixis | |||||
| 4 | x | 2.54 ± 0.01 | 7.51 ± 0.37 | 4x | 12x | 137 | 4x | 4x/2 | 2x | 2 | 2m:1p | 2.98 | Pseudogamous Apomixis | ||||
| 4x | 2.55 ± 0.04 | 7.04 ± 0.25 | 4x | 11x | 22 | 4x | 4x/2 | 3x | 1 | 2.7m:1p | 2.75 | Pseudogamous Apomixis | |||||
| 4x | 2.58 ± 0.12 | 5.83 ± 0.28 | 4x | 9x | 11 | 4x | 4x/2 | 1x | 1 | 8m:1p | 2.17 | Pseudogamous Apomixis | |||||
| 4x | 2.53 ± 0.05 | 5.08 ± 0.17 | 4x | 8x | 32 | 4x | 4x/2 | - | 0 | 4m:0p | 4:1 | Autonomous Apomixis | |||||
| 4x | 2.55 ± 0.08 | 4.46 ± 0.17 | 4x | 7x | 9 | 4x | Not resolved | ||||||||||
| 4x | 1.28 ± 0.00 | 3.84 ± 0.04 | 2x | 6x | 2 | 2x | 2x/2 | 1x or 2x | 2 or 1 | Haploid Parthenogenesis | |||||||
| 591 | |||||||||||||||||
a Maternal ploidy inferred from cytometric leaf measurements. b Embryo ploidy was increased compared to mother ploidy. c Biii hybrids. d Embryo ploidy was decreased compared to mother ploidy.
3.1. Seed Origin in Diploids
Diploids yielded exclusively sexually originated seeds having the same or increased ploidy level compared to the mother plant, which depended on sperm cell ploidies (Figure 2(S1,S2), Table 3). The profile 2x emb.:3x end. was as expected the most represented (103 seeds) and included both reduced female and male gametes. The profile 3x emb.:4x end. (eight seeds) was a result of interploid crosses between diploids and tetraploids. Those seeds originated from joint of reduced gametes (2x) from tetraploid males and a reduced egg cell (1x) of diploid mother.
3.2. Seed Origin in Triploids
All three analyzed triploid seeds had different profiles (Figure 2(S3,A1,A2), Table 3). The only sexual profile 3x emb.:4x end. from one seed included 2x male gamete and reduced (1x) egg cell (Table 3). The other two seeds were of apomictic origin. One included unreduced egg cell fecundated with reduced (1x) sperm cell creating 3x emb.:7x end. Profile, and the other included unreduced (3x) sperm cell creating 3x emb.:9x end. profile (Figure 2(A1,A2), Table 3).
4. Genotypic Variation in Cotoneaster integerrimus Populations
Five microsatellite loci were successfully amplified in the analyzed C. integerrimus populations and yielded a total of 99 alleles from 254 individuals (Table S2 could be found in https://www.mdpi.com/2223-7747/10/12/2798#supplementary).
A total of 92 multilocus genotypes (MLGs) were obtained from five loci (Ng, Table 4). Each of the studied diploids (N = 13) had a unique genotype (Table S2). The clonal genotypes (N = 32) were found within a polyploid pool of 241 individuals (Table S2). Each population contained at least one clonal genotype shared by a different number of individuals within a population, indicating clonal reproduction (Ni = 194, Table 4). The proportion of the detected clones (Ng/N) ranged from 0.1 to 0.91 (Table 4), reflecting the different level of clonal structure of populations. In each population, at least two individuals shared the same genotype (Table 4). Moreover, certain populations were completely clonal (Pu–Mt. Trebević, Si-Sinjajevina, He–Austria; Ng = 1, Table 4). In the majority of populations, the effective number of genotypes was higher than one, indicating the presence of unique genotypes in populations (Table 4). Different values of genotypic diversity of populations (Table 4) showed their diverse structure of clonal and unique genotypes, suggesting different ratios of sexuality and asexuality in populations.
Table 4. Genetic diversity measures for the analyzed Cotoneaster integerrimus populations derived from nuclear microsatellites.
| Locality (Site ID) | Number of Individuals Sampled ( | N | ) | Number of Multilocus Genotypes Detected ( | Ng | ) | Total Number of Individuals Belonging to a Clone ( | Ni | ) | Total Number of Unique Genotypes per Population ( | Un | ) | Effective Number of Genotypes ( | Eff | ) | Proportion of Clones Detected ( | Ng/N | ) | Genotypic Diversity ( | Div | ) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gornja Grkarica (Go) | 19 | 3 | 18 | 1 | 2.21 | 0.15 | 0.57 | |||||||||||||||||||||
| 43.74528 | 1.95 ± 0.05 | 2x | 1350 | -, -, - | -, -, - | 20, 98, 19 | 3x | 20, 98, 19 | ||||||||||||||||||||
| 103 | 1x | 1x/2 | 1x | 1 | 2m:1p | 1.52 | ||||||||||||||||||||||
| Umoljani (Um) | Sexual | 24 | 21 | 2 | Um | Umoljani, Mt. Bjelašnica, B&H | 18.22167 | 43.66194 | 1300 | 18, 108, 12 | 4, 3, 4 | 8, 27, 8 | 30, 138, 24 | |||||||||||||||
| 4 | 19 | 18 | 0.91 | 0.98 | 2x | 1.91 ± 0.05 | 2.59 ± 0.06 | 3x | 4x | 8 | b | 1x | 1x/2 | 2x | 1 | 1m:1p | 1.35 | Sexual | 3 | Vr | Vrbe, Čemerno, B&H | 18.57167 | 43.17861 | 1090 | -, -, - | 1, -, - | 19, -, 18 | 20, -, 18 |
| 3x | 1.94 | 2.64 | ||||||||||||||||||||||||||
| Vrbe (Vr) | 18 | 7 | 12 | 6 | 4.37 | 0.39 | 0.81 | 3x | 4x | 1 | 1x | 1x/2 | 2x | 1 | 1m:1p | |||||||||||||
| Bosiljna (Bo) | 16 | 5 | 14 | 5 | 3.55 | 0.31 | 1.5 | 0.76 | Sexual | 4 | Bo | Bosiljna, Mt. Čvrsnica, B&H | 17.48806 | 43.51194 | 1319 | -, -, - | -, -, - | 20, 7, 16 | 20, 7, 16 | |||||||||
| 3x | 1.85 | 3.93 | 3x | 7x | 1 | 3x | 3x/2 | 1x | 1 | 6m:1p | 1.35 | Pseudogamous Apomixis | ||||||||||||||||
| Rujište (Ru) | 19 | 4 | 17 | 4 | 2.39 | 0.21 | 0.61 | 5 | Ru | |||||||||||||||||||
| Borova glava (Bg) | 21 | Rujište, Mt. Prenj, B&H | 17.95472 | 43.46194 | 1010 | -, -, - | 1, -, 1 | 17, 14, 18 | 18,14, 19 | |||||||||||||||||||
| 2 | 20 | 1 | 1.09 | 0.1 | 0.09 | 6 | Bg | Borova glava, Livno, B&H | 17.12139 | 43.77861 | 1183 | 1, 3, - | -, -, - | 22, 28, 21 | 23, 31, 21 | |||||||||||||
| Sovićka vrata (So) | 14 | 6 | 11 | 4 | 4.07 | 0.42 | 0.81 | 7 | So | Sovićka vrata, Blidinje, B&H | 17.50472 | 43.59528 | 1220 | -, -, - | 1, -, 1 | |||||||||||||
| Puhova ravan (Pu) | 20 | 1 | 20 | 1 | 1 | 16, 4, 13 | 0.05 | 17, 4, 14 | ||||||||||||||||||||
| 0 | 8 | Pu | Puhova ravan, Mt. Trebević, B&H | 18.43833 | 43.82861 | 1450 | -, -, - | -, -, - | 20, 55, 20 | 20, 55, 20 | ||||||||||||||||||
| 9 | ||||||||||||||||||||||||||||
| Čavljak (Ca) | 19 | 5 | 16 | 4 | 1.57 | 0.26 | 0.38 | Ca | Čavljak, Mt. Ozren, B&H | 18.43833 | 43.91194 | 1237 | -, -, - | -, -, - | 21, 75, 19 | 21, 75, 19 | ||||||||||||
| Devečanske stijene (De) | 19 | 6 | 10 | De | Devečanske stijene, Mt. Vlašić, B&H | 17.62139 | 44.27889 | 1752 | 1, 2, 1 | -, -, - | 18, 57, 18 | 19, 59, 19 | ||||||||||||||||
| 16 | 11 | Rt | Mt. Rtanj, Serbia | 21.88917 | 43.77861 | 1282 | -, -, - | 1 *, -, 1 | 8 *, -, 8 | 9, -, 9 | ||||||||||||||||||
| 12 | Mu | Mt. Mučanj, Serbia | 20.03889 | 43.54528 | 1530 | -, -, - | -, -, - | 7 *, 36, 7 | 7, 36, 7 | |||||||||||||||||||
| 13 | Pr | Premužićeva staza, Mt. Velebit, Croatia | 14.98722 | 44.76222 | 1613 | -, -, - | 1 *, -, 1 | 3 *, 2, 3 | 4, 2, 4 | |||||||||||||||||||
| 6 | 2.63 | 0.31 | 0.65 | |||||||||||||||||||||||||
| Mt. Rtanj (Rt) | 9 | 4 | 7 | 4 | 3 | 0.44 | 0.75 | |||||||||||||||||||||
| Mt. Mučanj (Mu) | 7 | 2 | 7 | 2 | 1.86 | 0.28 | 0.47 | |||||||||||||||||||||
| Premužićeva staza (Pr) | 4 | 3 | 2 | 3 | 2.66 | 1 | 0.83 | |||||||||||||||||||||
| Vošac (Vo) | 9 | 4 | 7 | 3 | 2.07 | 0.33 | 14 | Vo | Vošac, Mt. Biokovo, Croatia | 17.05472 | 43.31194 | 1320 | -, -, - | 1 *, -, 1 | 8 *, 58, 8 | 9, 58, 9 | ||||||||||||
| 0.58 | ||||||||||||||||||||||||||||
| Mt. Sinjajevina (Si) | 5 | 1 | 5 | 1 | 1 | 0.20 | 0 | 15 | Si | Mt. Sinjajevina, Monte Negro | 19.33861 | 42.96167 | 1773 | -, -, - | -, -, - | 5 *, -, 5 | 5, -, 5 | |||||||||||
| Hechenberg-Ostgipfel (He) | 5 | 1 | 5 | 1 | 1 | 0.20 | 0 | 16 | He | Hechenberg-Ostgipfel von Kranebitten, Innsbruck, Austria | 11.31972 | 47.27972 | 1390 | -, -, - | -, -, - | 5 *, -, 5 | 5, -, 5 | |||||||||||
| Rotenfels (Ro) | 5 | 3 | 3 | 3 | 2.27 | 0.60 | 0.70 | 17 | Ro | Rotenfels, Rheinland-Pfalz, Germany | 7.818611 | 49.83028 | 232 | -, -, - | -, -, - | 5 *, 6, 5 | ||||||||||||
| Monte Ventosola (Mo) | 4 | 3 | 2 | 3 | 2.66 | 0.75 | 5, 6, 5 | |||||||||||||||||||||
| 0.83 | 18 | Mo | Monte Ventosola, Monte Sibillini, Umbria, Italy | 13.17028 | 42.77833 | 1493 | -, -, - | -, -, - | 4 *, 8, 4 | 4, 8, 4 | ||||||||||||||||||
| Mt. Suva Gora (Su) | 7 | 19 | Su | Mt. Suva Gora, Northern Macedonia | 21.25583 | 41.84472 | 1184 | -, -, - | -, -, - | 7 *, -, 7 | 7, -, 7 | |||||||||||||||||
| 20 | Ol | Mt. Olympus, Greece | 22.37278 | 40.07778 | 1935 | -, -, - | -, -, - | 5 *, -, 5 | 5, -, 5 | |||||||||||||||||||
| 21 | Pa | Mt. Pangaion, Greece | 24.07333 | 40.92778 | 1662 | -, -, - | -, -, - | 5 *, -, 5 | 5, -, 5 | |||||||||||||||||||
| Total | N | per cytotype | 20, 113, 13 | 10, 3, 9 | 243, 475, 232 | 273, 591, 254 | ||||||||||||||||||||||
* Ploidy level was inferred from microsatellite data.
Geographic distribution of cytotypes showed a clear prevalence of tetraploids (89% in the total sample) in each population except the Umoljani (Um) in Bosnia and Herzegovina (Table 2, Figure 1B). The sample for Figure 1B encompassed the ploidies obtained from flow cytometry as well as the estimated ploidy levels from microsatellite data for 65 individuals (explained in Table 2 and the Material and Methods section). Diploids (7.3% in total sample) were recorded in only three populations with a single individual in Borova glava (Bg) and Devečani (De), and 18 individuals in Umoljani (Um) (Table 2, Figure 1B). Triploids (3.7% in total sample) were observed in seven populations and were represented with just one individual in six populations (Vrbe-Vr, Rujište-Ru, Sovićka vrata-So, Vošac-Vo, Premužićeva staza-Pr, Rtanj-Rt) and four individuals in the population Umoljani (Table 2).
3. Flow Cytometric Seed Screening
Only unambiguous results of flow cytometric seed screening (FCSS) are presented. Approximately one quarter of analyzed seeds had minor additional peaks corresponding to doubled nuclear DNA levels mirroring endoreplicated embryo and/or endosperm nuclei. In total, the reproductive mode was characterized for 591 seeds from three C. integerrimus cytotypes (Table 2).
In general, inferred pathways of sexual and asexual seed formation showed great diversity and included both reduced and unreduced gametes within each cytotype (Figure 2, Table 3). The tetraploid cytotype was the most abundant, and it consequently resulted in the highest number of reproductive pathways, predominantly apomictic. Diploids involved only sexual reproduction (Table 3). The triploid cytotype was represented with only three seeds, one of sexual and two of apomictic origin (Table 3). In any case, sexual and asexual seed formation was clearly distinguished.
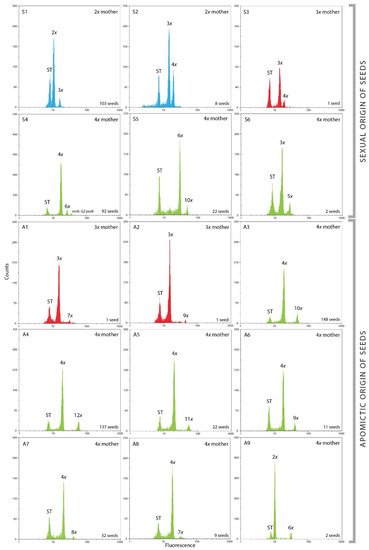

Figure 2. Flow cytometric histograms (log abscissa) of Cotoneaster integerrimus seed formation from sexual (S1–S6) or asexual (A1–A9) origin. The first fluorescence peaks correspond to the internal standard (ST, Oryza sativa L. ssp. japonica ‘Nipponbare’), the second ones to the embryo, and the third ones to the endosperm (blue: 2x mother, red: 3x mother, green: 4x mother).
Table 3. Flow cytometric seed screening results and hypothesized pathways of seed formation and reproduction modes in analyzed Cotoneaster integerrimus populations.
| 3x | |||||||
| 1.94 | |||||||
| 5.72 | 3x | 9x | |||||
| 3 | |||||||
| 4 | |||||||
| 2 | |||||||
| 1.81 | |||||||
| 0.71 | |||||||
| 0.52 | |||||||
| Mt. Olympus (Ol) | 5 | 4 | 2 | 4 | 3.57 | 0.8 | 0.90 |
| Mt. Pangaion (Pa) | 5 | 4 | 2 | 4 | 3.57 | 0.8 | 0.90 |
| 254 | 92 | 194 | 81 |
While the majority of clonal genotypes (N = 29) were geographically restricted within a single or two populations, the two clonal genotypes occurred at multiple sites (Table S2). The more widespread clonal genotype occurred in populations Bo–Borova glava (N = 20), Go–Mt. Igman (N = 9), Ru–Rujište (N = 9), So–Sovićka vrata (N = 2), and Um–Umoljani (N = 1), which was equivalent to 17% of all polyploids. The second clonal genotype was less widespread and was shared by plants from three populations: So–Sovićka vrata (N = 4), Um–Umoljani (N = 1), and Vr–Vrbe (N = 3). One hundred and thirty-seven (56.8%) individuals belonged to 25 spatially limited clonal genotypes, while other individuals possessed unique genotypes.
Principal coordinate analysis (PCoA) based on Jaccard distances was applied on C. integerrimus polyploid MLGs (Figure 3). PCoA revealed the pattern of grouping of MLGs dispersed along the first (PCo1) and the second (PCo2) coordinates (Figure 3). A weak regional geographic pattern of separation among MLGs groups was evident. The MLGs from western Balkans formed three groups along the PCo1: the first group consisted of MLGs from Bosnia and Herzegovina (Bg, Bo, Ru, Go, So, Um, Vr) and Croatia (Vo); the second consisted of several Bosnian MLGs (Go and Pu); and the third one was the most heterogeneous and consisted of MLGs originating from Bosnia and Herzegovina (Ca, De, Pr, Um, Vr), Croatia (Pr), and Montenegro (Si). MLGs from Italy (Mo), Austria (He), and Germany (Ro) were intermingled with Balkan MLGs. The southern Balkan MLGs from Greece (Ol and Pa) and North Macedonia (Su) clustered into a separate group along the second coordinate (Figure 3). Serbian MLGs (Mu and Rt) formed a group neighboring the southern Balkan cluster. Certain MLGs from De and Um populations were interspersed within the southern Balkan cluster.
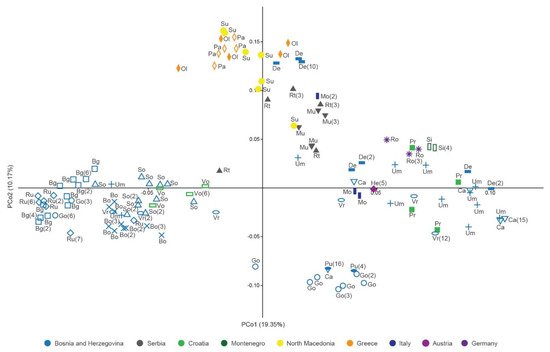

Figure 3. Principal coordinate analysis of the Jaccard distance matrix of multilocus genotypes of Cotoneaster integerrimus polyploids according to nuclear microsatellite data. Population abbreviations correspond to those in the Table 4. The numbers in the brackets denote the number of clones.
References
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome size diversity and its impact on the evolution of land plants. Genes 2018, 9, 88.
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424.
- Soltis, D.E.; Visger, C.J.; Soltis, P.S. The polyploidy revolution then and now: Stebbins revisited. Am. J. Bot. 2014, 101, 1057–1078.
- Ďurkovič, J.; Kardošová, M.; Čaňová, I.; Lagaňa, R.; Priwitzer, T.; Chorvát, D.; Cicák, A.; Pichler, V. Leaf traits in parental and hybrid species of Sorbus (Rosaceae). Am. J. Bot. 2012, 99, 1489–1500.
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; Depamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348.
- Hegarty, M.J.; Hiscock, S.J. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 2008, 18, 435–444.
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17.
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016, 210, 391–398.
- Dobeš, C.; Scheffknecht, S.; Fenko, Y.; Prohaska, D.; Sykora, C.; Hülber, K. Asymmetric reproductive interference: The consequences of cross-pollination on reproductive success in sexual–apomictic populations of Potentilla puberula (Rosaceae). Ecol. Evol. 2019, 8, 365–381.
- Hörandl, E. The evolution of self-fertility in apomictic plants. Sex. Plant Reprod. 2010, 23, 78–86.
- Barrett, S.C.H. The evolution, maintenance, and loss of self-incompatibility systems. In Plant Reproductive Ecology, 1st ed.; Lovett, J.D., Lovett, L.D., Eds.; Oxford University Press: New York, NY, USA, 1988; pp. 98–124.
- Hörandl, E. The complex causality of geographical parthenogenesis. New Phytol. 2006, 171, 525–538.
- Hojsgaard, D.; Hörandl, E. The rise of apomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358.
- Hörandl, E.; Cosendai, A.-C.; Temsch, E.M. Understanding the geographic distributions of apomictic plants: A case for a pluralistic approach. Plant Ecol. Divers. 2008, 1, 309–320.
- Ozias-Akins, P.; van Dijk, P.J. Mendelian genetics of apomixis in plants. Ann. Rev. Genet. 2007, 41, 509–537.
- Mráz, P.; Mrázová, V. Greater reproductive assurance of asexual plant compared to sexual relative in a low density sympatric population–experimental evidence for pollen limitation. J. Evol. Biol. 2021, 34, 1503–1509.
- Alonso-Marcos, H.; Nardi, F.D.; Scheffknecht, S.; Tribsch, A.; Hülber, K.; Dobeš, C. Difference in reproductive mode rather than ploidy explains niche differentiation in sympatric sexual and apomictic populations of Potentilla puberula. Ecol. Evol. 2019, 9, 3588–3598.
- Coughlan, J.M.; Han, S.; Stefanović, S.; Dickinson, T.A. Widespread generalist clones are associated with range and niche expansion in allopolyploids of Pacific Northwest Hawthorns (Crataegus L.). Mol. Ecol. 2017, 26, 5484–5499.
- Hojsgaard, D.; Hörandl, E. A little bit of sex matters for genome evolution in asexual plants. Front. Plant Sci. 2015, 6, 82.
- Majeský, L.; Vašut, R.J.; Kitner, M.; Trávníček, B. The pattern of genetic variability in apomictic clones of Taraxacum officinale indicates the alternation of asexual and sexual histories of apomicts. PLoS ONE 2012, 7, e41868.
- Paun, O.; Stuessy, T.F.; Hörandl, E. The role of hybridization, polyploidization and glaciation in the origin and evolution of Ranunculus cassubicus complex. New Phytol. 2006, 171, 223–236.
- Lepší, M.; Koutecký, P.; Nosková, J.; Lepší, P.; Urfus, T.; Rich, T.C.G. Versatility of reproductive modes and ploidy level interactions in Sorbus s.l. (Malinae, Rosaceae). Bot. J. Linn. Soc. 2019, 191, 502–522.
- Hajrudinović, A.; Siljak-Yakovlev, S.; Brown, S.C.; Pustahija, F.; Bourge, M.; Ballian, D.; Bogunić, F. When sexual meets apomict–genome size, ploidy level and variation of reproductive mode in Sorbus aria s.l. and S. austriaca (Rosaceae) in Bosnia and Herzegovina. Ann. Bot. 2015, 116, 301–312.
- Potter, D.; Eriksson, T.; Evans, R.C.; Oh, S.H.; Smedmark, J.E.E.; Morgan, D.R.; Kerr, M.; Robertson, K.R.; Arsenault, M.P.; Dickinson, T.A.; et al. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 2007, 266, 5–43.
- Fryer, J.; Hylmö, B. Cotoneasters: A Comprehensive Guide to Shrubs for Flowers, Fruit, and Foliage, 1st ed.; Timber Press: London, UK, 2009; p. 334.
- Dickoré, W.B.; Kasperek, G. Species of Cotoneaster (Rosaceae, Maloideae) indigenous to, naturalising or commonly cultivated in Central Europe. Willdenowia 2010, 40, 13–45.
- Lu, L.D.; Brach, A.R. Cotoneaster. In Flora of China, 1st ed.; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2003; Volume 9, pp. 85–108.
- Meng, K.K.; Chen, S.-F.; Xu, K.-W.; Zhou, R.-C.; Li, M.-W.; Dhamala, M.K.; Liao, W.-B.; Fan, Q. Phylogenomic analyses based on genome-skimming data reveal cyto-nuclear discordance in the evolutionary history of Cotoneaster (Rosaceae). Mol. Phylogenetics Evol. 2021, 158, 107083.
- Bourge, M.; Mahmutović, I.; Hajrudinović-Bogunić, A.; Siljak-Yakovlev, S.; Silajdžić, E.; Brown, S.C.; Muratović, E.; Bogunić, F. Cytotypes and reproductive modes of Cotoneaster integerrimus (Rosaceae) from Bosnia and Herzegovina: Preliminary results. In Proceedings of the Cytometrie: 19e Congrès annuel de l’association Française de Cytométrie, Antibes, France, 18–20 November 2015; p. 285.
- Macková, L.; Nosková, J.; Ďurišová, Ľ.; Urfus, T. Insights into the cytotype and reproductive puzzle of Cotoneaster integerrimus in the Western Carpathians. Plant Syst. Evol. 2020, 306, 58.
- Li, M.; Chen, S.; Zhou, R.; Fan, Q.; Li, F.; Liao, W. Molecular evidence for natural hybridization between Cotoneaster dielsianus and C. glaucophyllus. Front. Plant Sci. 2017, 8, 704.
- Rothleutner, J.R.; Friddle, M.W.; Contreras, R.N. Ploidy levels, relative genome sizes, and base pair composition in Cotoneaster. J. Amer. Soc. Hort. Sci. 2016, 141, 457–466.
- Nybom, H.; Bartish, I.V. DNA markers and morphometry reveal multiclonal and poorly defined taxa in an apomictic Cotoneaster species complex. Taxon 2007, 56, 119–128.
- Kroon, G.H. Polyploidy in Cotoneaster II. Acta Bot. Neerl. 1975, 24, 417–420.
- Sax, H.J. Polyploidy and apomixis in Cotoneaster. J. Arnold Arbor. 1954, 35, 334–365.
- Hjelmquist, H. Embryo sac development of Cotoneaster. Bot. Not. 1962, 115, 209–236.
- Mandryk, V.Y. Seed reproduction features in Cotoneaster melanocarpus Fisch. ex Blytt. (Rosaceae) from Ukrainian Carpathian flora. Ukr. Botan. Zhurn. 2006, 19, 66–72. (In Ukrainian)
- Mandryk, V.Y. Embryologic investigation of Cotoneaster melanocarpus Fisch. ex Blytt (Rosaceae). Ukr. Botan. Zhurn. 1994, 51, 86–93. (In Ukrainian)
- Flinck, K.E.; Hylmö, B. Cotoneaster harrysmithii, a new species from western China. Bot. Not. 1962, 112, 17–64.
- Kšiňan, S.; Ďurišová, L.; Elias, P. Genome size estimation of Cotoneaster species (Rosaceae) from the Western Carpathians. Biologia 2021, 76, 1–12.
- Doležel, J.; Bartoš, J.; Voglmayr, H.; Greilhuber, J. Nuclear DNA content and genome size of trout and human. Cytometry 2003, 51, 127–128.
More
