Over time, machine learning (ML), a component of artificial intelligence (AI), has been implemented in a variety of medical specialties, such as radiology, pathology, gastroenterology, neurology, obstetrics and gynecology, ophthalmology, and orthopedics, with the goal of improving the quality of healthcare and medical diagnosis.
In clinical gastroenterology practice, due to technological developments, estimates show that AI could have the ability to create a predictive model; for instance, it could develop an ML model that can stratify the risk in patients with upper gastrointestinal bleeding, establish the existence of a specific gastrointestinal disease, define the best treatment, and offer prognosis and prediction of the therapeutic response. In this context, by applying ML or deep learning (DL) (AI using neural networks), clinical management in gastroenterology can begin to focus on more personalized treatment centered on the patient and based on making the best individual decisions, instead of relying mostly on guidelines developed for a specific condition. Moreover, the goal of implementing these AI-based algorithms is to increase the possibility of diagnosing a gastrointestinal disease at early stage or the ability to predict the development of a particular condition in advance.
Because both AI and gastroenterology encompass many subdomains, the interaction between them might take on various forms. In recent years, we have witnessed a large explosion of research in attempts to improve various fields of gastroenterology, such as endoscopy, hepatology, inflammatory bowel diseases, and many others, with the aid of ML. We also note that, because of the requirement to diagnose more patients with gastrointestinal cancers at an early stage of the disease, which is associated with curative treatment and better prognosis, many studies were developed to address improvement of the detection of these tumors with the aid of AI.
The term ML, introduced for the first time in 1959 by Arthur Samuel from the IBM company, refers to an IT domain whereby a computer system can acquire the ability to “learn” by using data without specific programming and can therefore develop a predictive mathematical algorithm based on input data, using recognition of “features”. The ML “model” is subsequently able to adapt to new situations in which it becomes able to predict and make decisions.
- artificial intelligence
- computer-assisted diagnosis
- machine learning
- gastroenterology
- endoscopy
1. Introduction
Three main types of learning methodologies are recognized, namely, supervised learning, in which the computer learns from familiar patterns; unsupervised learning, in which the computer discovers the common aspects in unknown patterns; and, finally, reinforcement learning, in which the computer has the ability to learn from trial and error[1][2] (Figure 1). Clustering algorithms are based on unsupervised learning, in which unlabeled data self organizes to predict outcomes (e.g., clustering). Classification and regression algorithms are based on unsupervised learning, in which prelabeled data train a model to predict new outcomes. Rewards and recommendations algorithms are based on reinforcement learning, which gives feedback to an algorithm when it does something right or wrong.
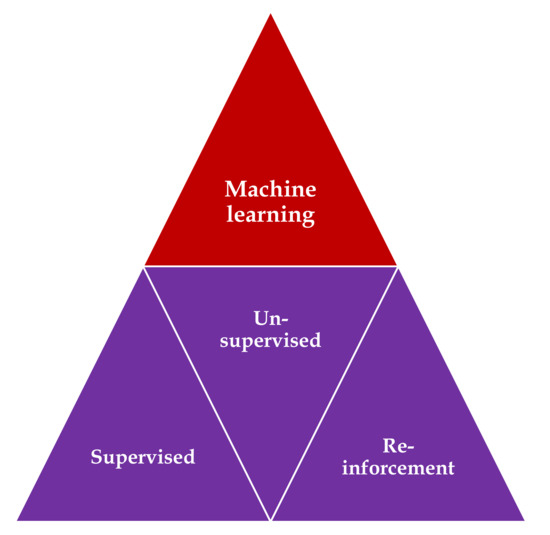
Figure 1. Types of machine learning algorithms: supervised learning—task driven (classification); unsupervised learning—data driven (clustering); and reinforcement learning—algorithm learns from trial and error.
The predictive models encompass the key elements of the “training”, “validation”, and “testing” datasets. Approximately 70% of samples are commonly used in the initial training set to develop the model, and the remaining 30% of the samples are used as model validation and testing sets, but these percentages may vary with the application[3].
AI was implemented in the medical field, using different types of ML, such as binary classifiers, Bayesian inferences, decision trees, ensemble trees, linear discriminants, support vector machines (SVM), k-nearest neighbors, logistic regression, and artificial neural networks (ANNs)[4][5].
Support vector machine (SVM), which was invented in 1963, before the development of DL[6], represents a supervised learning model, a discriminative algorithm that uses a dividing hyperplane. SVM demonstrated its best accuracy in classification and regression analysis.
2.ML Using Hand-Crafted Features (Conventional Algorithms)
For a long time, ML using images (in the field of gastroenterology, we are using endoscopic images) relied primarily on hand-crafted features. In this context, IT specialists coded a mathematical description of specific patterns, such as color and texture. The researchers manually indicated the potential features of the images based on clinical expertise. A classifier was trained to distinguish between different classes of features, and eventually, the model was able to use this knowledge to recognize the class in a new set of images[2].
3. ML Using Deep Learning (DL)
DL refers to a subset of ML techniques that is built from multiple-layered neural network algorithms. They represent ML algorithms that use layers of nonlinear processing for “feature extraction”, which is the selection of powerful predictive variables, and “transformation”, which refers to changing the data for more efficient construction of the model[7].
3.1. Neural Networks
Neural networks represent a specific area of ML that shows similarities with the human brain, namely densely interconnected neurons, with the aim of recognizing specific patterns, extracting features, or learning different characteristics of the training dataset to elaborate a concrete result[3][7].
Therefore, in the case of an artificial neural network (ANN), we use a fully connected neural network in which the outputs of the neurons of one layer represent the input for the neurons of the next layer. Each connection has a specific weight that is learned during the training process, and the model is based on a nonlinear sigmoidal function.
A deep neural network represents an ANN containing several hidden layers between the input and output layer. This technology proved to possess excellent accuracy for establishing diagnosis and predicting prognosis in the medical area. In most cases, DL outperforms the hand-crafted algorithm, but it requires a larger quantity of data for learning[8][9]. Fortunately, most of the initial weaknesses and limits of the deep neural network have been overcome by the recent availability of big data for training and the major progress in computing software power[4][5].
One drawback of DL is its “black-box” nature, meaning that the system cannot apply reason to the machine-generated decision, which can be a confusing aspect for the endoscopist. However, a new research area known as “interpretable DL” has attracted recent attention through its attempt to present an argument-based framework for decision-making[10]. Although DL models proved to be the most performant algorithms in fitting the data, one of their limits is their dependency on the training dataset. This “overfitting” error appears if the training database is not sufficiently diverse or contains bias. In that case, the results might not be validated and implemented in real-life circumstances. To enlarge the training datasets, these approaches might include images showing normal aspects and images containing pathologic lesions. Additionally, most of the recent studies use augmentation of the image-based data by resizing and cropping of the frame, with a subsequent flipping along the axis[5][11].
Convolutional neural networks (CNN) represent a specific class of ANN composed of convolutional and pooling layers with the role of extracting specific features and fully connected layers that fulfill the task of elaborating the definitive classification.
The input images are subjected to the preprocessing procedure of filtering (convolution) to extract specific features. Subsequently, the convolution filter undergoes a learning process to elaborate the performant feature maps, which are compressed to smaller sizes. At the end, the fully connected layers combine the selected features for design of the final model. In case of CNN, the number of weights is significantly lower than that of the fully connected networks. The CNN has demonstrated excellent performance in image analysis (Figure 2).
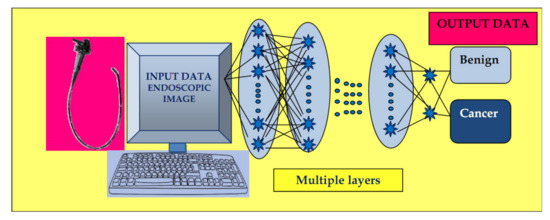
Figure 2. Convolutional neural network (CNN) system: input layer with raw data of the endoscopic image, multiple layers with the role of extracting specific features, and elaborating image classification in the output layer.
The concept of CNN was developed independently by several different groups during the 1970s and 1980s. The proof of concept of CNN emerged in the late 1980s when Bengio, Hinton, and LeCun started to exchange ideas in this field, and the first paper on backpropagation procedure on CNN was published in 1990[12]. In 1998, LeCun wrote an overview paper on the principles of training of deep neural networks using gradient-based optimization, showing that CNN can be combined with search or inference mechanisms to model interdependent complex outputs[13]. In 2006, the Canadian Institute for Advanced Research (CIFAR) revived the interest in deep feedforward networks by connecting a group of researchers who introduced unsupervised learning procedures. The first major application of this approach was in speech recognition, which was developed during the following years; by 2012, new versions were already being deployed in Android phones. Since the 2000s, CNN have been successfully applied to the detection, recognition, and segmentation of objects/regions in images; a major recent practical success of CNN is face recognition. Despite these advances, CNN was neglected by most of the computer-vision communities until the ImageNet competition (2012)[14].
In recent years, we have observed the impressive emergence of complex CNNs constructed from more than 100 layers, mostly due to an increased interest in this field and initiation of a great number of scientific activities. Due to the annual software contest known as the ImageNet Large Scale Visual Recognition Challenge (ILSVRC), designated for creation of AI, a great variety of software programs have been developed, such as residual nets (e.g., AlexNet, GoogleNet, InceptionResNet, and ResNet from Microsoft and many other variants), fully convolutional networks (FCN), U-Net (which is based on an encoder–decoder mechanism for pixelwise classification and is mostly used in segmentation processing on test images[15][16][17]), and others. As foreseen by LeCun[14], human vision, natural language understanding, and major progress in AI will materialize by using systems that combine representation learning with complex reasoning.
In the area of gastroenterology, which is overwhelmed by a notably large amount of clinical data and endoscopic or ultrasound images, this technology has been applied to aid clinicians in establishing diagnosis, estimating prognosis, and analyzing images.
3.2. Computer Vision
Computer vision refers to the specific use of computer systems in the processing of images/videos, and the possibility of acquiring information from this processing. We must note that a multitude of technological developments have been demonstrated recently in this domain. In the medical field, clinicians work with large amounts of visual data that must be analyzed to elaborate the proper diagnosis and choice of the best treatment, especially in domains such as radiology or endoscopy. In the latter domain, CNNs were elaborated for different purposes such as esophageal/gastric cancer detection[18][19] and “real-time” polyp detection/differentiation between polyp types[20][21], among others.
References
- Topol, E. Deep Medicine. Hachette Book Group USA, 2019; pp. 17-24
- Ebigbo, A.; Palm, C.; Probst, A; Mendel, R.; Manzeneder, J.; Prinz, F.; de Souza, L.A.; Papa, J.P.; Siersema, P.; Messmann, H. A technical review of artificial intelligence as applied to gastrointestinal endoscopy: clarifying the terminology. Endosc Int Open. 2019, 7(12), E1616-E1623. [DOI: 10.1055/a-1010-5705. PMID: 31788542]
- Ruffle, J.K.; Farmer, A.D.; Aziz, Q. Artificial intelligence-assisted gastroenterology- promises and pitfalls. Am J Gastroenterol. 2019, 114(3), 422-428. [PMID: 30315284 DOI: 10.1038/s41395-018-0268-4]
- Dey, A. Machine learning algorithms: a review. IJCSIT. 2016, 7, 1174–9.
- Yang, Y.J.; Bang, C.S. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019, 25(14), 1666-1683. [DOI: 10.3748/wjg.v25.i14.1666 PMID: 31011253]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the fifth annual workshop on Computational learning theory (COLT ’92). Association for Computing Machinery, New York, NY, USA, 1992; 144–152. [DOI:https://doi.org/10.1145/130385.130401]
- Schmidhuber, J. Deep learning in neural networks: an overview. Neural Netw. 2015, 61, 85–117. [DOI: 10.1016/j.neunet.2014.09.003 PMID: 25462637]
- Mori, Y.; Kudo, S.; Mohmed, H.; Misawa, M.; Ogata, N.; Itoh, H.; Oda, M.; Mori, K. Artificial intelligence and upper gastrointestinal endoscopy: Current status and future perspective. Digestive Endoscopy 2019, 31, 378–388. [DOI 10.1111/den.13317]
- Khan, S.; Yong, S. A comparison of deep learning and handcrafted features in medical image modality classification. 2016 3rd International Conference on Computer and Information Sciences (ICCOINS), Kuala Lumpur, Malaysia, 2016, pp. 633-638. [DOI: 10.1109/ICCOINS.2016.7783289]
- Philbrick, K.A.; Yoshida, K.; Inoue, D; Akkus, Z.; Kline, T.L; Weston, A.D.; Korfiatis, P.; Takahashi, N.; Erickson, B.J. What does deep learning see? Insights from a classifier trained to predict contrast enhancement phase from CT images. Am. J. Roentgenol. 2018, 211, 1184–93. [DOI: 10.2214/AJR.18.20331 PMID: 30403527]
- Le Berre, C.; Sandborn, W.J.; Aridhi, S.; Devignes, M-D.; Fournier, L.; Smaïl-Tabbone, M.; Danese, S.; Peyrin-Biroulet, L. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology 2020, 158, 76–94. [PMID: 31593701 DOI: 10.1053/j.gastro.2019.08.058]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Handwritten digit recognition with a back-propagation network. In: Proc. Advances in Neural Information Processing Systems 1990; 396–404
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998; 86, 2278–2324 [DOI: 10.1109/5.726791]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436-444 [PMID: 26017442 DOI:10.1038/nature14539]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, USA, 2016; 770–778.
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition, Boston, USA, 2015; 3431-3440.
- Ronneberger, O.; Fischer.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention, Munich, Germany, 2015; 234–241. [DOI https://doi.org/10.1007/978-3-319-24574-4_28]
- Ebigbo, A.; Mendel, R.; Probst, A.; Manzeneder, J.; Prinz, F.; de Souza, L.Jr.; Papa, J.; Palm, C.; Messman, H. Real- time use of artificial intelligence in the evaluation of cancer in Barrett’s oesophagus. Gut 2020, 69, 615–616. [DOI:10.1136/gutjnl-2019-319460]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; Tada, T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018, 21(4), 653-660. [DOI: 10.1007/s10120-018-0793-2. PMID: 29335825]
- East, J.E.; Rees, C.J. Making optical biopsy a clinical reality in colonoscopy. Lancet Gastroenterol Hepatol. 2018, 3(1), 10-12. [DOI: 10.1016/S2468-1253(17)30366-7. PMID: 29254613]
- Yuan, Y.; Meng, M.Q. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017, 44, 1379–89. [DOI: 10.1002/mp.12147. PMID: 28160514]
