The spatial organization of the genome into the cell nucleus plays a central role in controlling several genome functions, such as gene expression and DNA replication timing during the S-phase of the cell cycle. Here we show how chromosomes are organized in the cell nucleus according to the gene density and to the GC-level of the various chromosomal bands, allowing a corrected and coordinated gene expression during cell life. The human genome, such as the genome of the other mammals, is composed by two very different parts: one very gene-dense, replicated at the onset of the S-phase, very GC-rich and the other endowed bywith opposite features. These two genomic compartments are localized in thefar apart within a chromosome distant one to each other, with regions having intermediate properties located between them. This determines a zig-zag organization of the larger chromosomes, to position the gene-poorest genome compartment at the nuclear periphery and the gene-richest one at the nuclear interior.
- cancer
- genome organization
- cell nucleus
- chromosomal bands
1. Radial nuclear organization of the chromosome territories
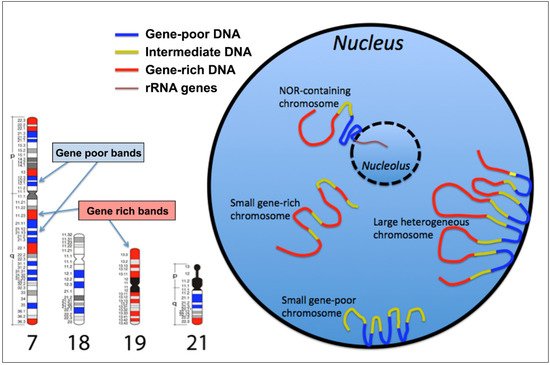
2. The GC-richest and the GC-poorest nuclear compartments
3. Gene location in the cell nucleus, and transcriptional activity
Since genes are part of chromosomes it also matters where those genes sit in relation to the body of their home interphase chromosome territory, i.e. intra-chromosomal organization. Indeed, gene loci can be located deep within chromosome territories, more towards the surface of the chromosome territories, or even at some distance from the main body of the chromosome territory, out on a chromatin loop. There is evidence that even within interphase chromosomes there is spatial organization of gene-rich and gene-poor areas, which has been revealed by FISH, with inactive genes more likely to be located in the interior of chromosome territories [23][26][27][28][29][30]. When chromosomes are located at the nuclear periphery, their active genes are generally pointed towards the nuclear interior and not at the nuclear envelope side [10][31].
The arrangement of genes within a chromosome territory would permit the active regions of the genome to be closer and exposed to the components, machinery and structures required for transcription and processing[32]. We know that active transcription can take place at the surfaces and around channels that inveigle their way into chromosomes[33]. However, some gene loci are located away from the main body of their chromosome territory out on loops to be transcribed at a distance from the chromosome[10][29][34][35], at transcription factories or splicing speckles[36][37], becoming colocalized with other genes[38][39]. This positioning of chromatin loops belonging to different chromosomes in the same nuclear compartments can determine chromosomal rearrangements such as translocations.
Several studies have explored the link between chromosomal abnormalities and their effect on gene expression [reviewed in [40]]. Due to the precise nature of chromosome positioning within the nucleus, according to the GC-level of the genomic regions composing each band, the gross changes created by chromosomal rearrangements lead to major spatial disorders that go beyond local and in-cis effects. Chromosomal anomalies determine a change in the nuclear location of the chromosomal bands located around the breakpoints, and this spatial repositioning can cause changes in the accessibility status of the chromatin involved in the rearrangement, with repercussions on the transcriptional regulation of genomic loci not directly altered by the rearrangements[41].
4. Conclusion
Many different experimental procedures have demonstrated the presence of two very different nuclear compartments, one endowed with a high GC level, a high gene density, a very early replication during the S-phase of the cell cycle, and a position in the inner part of the nucleus. Another compartment, generally located more peripherally in the nucleus, is endowed with opposite properties. Rearrangements between loci belonging to these two different compartments determines the repositioning of genes in a compartment with different environmental properties, this determining the ectopic activation or inactivation of the relocated genes.References
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301.
- Boyle, S.; Gilchrist, S.; Bridger, J.M.; Mahy, N.L.; Ellis, J.A.; Bickmore, W.A. The spatial organization of human chromo-somes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 2001, 10, 211-219.
- Croft, J.; Bridger, J.M.; Boyle, S.; Perry, P.; Teague, P.; Bickmore, W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999, 145, 1119-1131.
- Malhas, A.; Lee, C.F.; Sanders, R.; Saunders, N.J.; Vaux, D.J. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 2007, 176, 593-603.
- Tanabe, H.; Mùller, S.; Neusser, M.; von Hase, J.; Calcagno, E.; Cremer, M.; Solovei, S.; Cremer, C.; Cremer, T. Evolution-ary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 4424-4429.
- Saccone, S.; De Sario, A.; Della Valle, G.; Bernardi, G. The highest gene concentrations in the human genome are in telo-meric bands of metaphase chromosomes. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 4913-4917.
- Saccone, S.; De Sario, A.; Wiegant, J.; Raap, A.K.; Della Valle, G.; Bernardi, G. Correlations between isochores and chro-mosomal bands in the human genome. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 11929-11933.
- Saccone, S.; Federico, C.; Solovei, I.; Croquette, M.F.; Della Valle, G.; Bernardi, G. Identification of the gene-richest bands in human prometaphase chromosomes. Chromosome Res. 1999, 7, 379-386.
- Federico, C.; Andreozzi, L.; Saccone, S.; Bernardi, G. Gene density in the Giemsa bands of human chromosomes. Chromo-some Res. 2000, 8, 737-746.
- Federico, C.; Cantarella, C.D.; Di Mare, P.; Tosi, S.; Saccone, S. The radial arrangement of the human chromosome 7 in the lymphocyte cell nucleus is associated with chromosomal band gene density. Chromosoma 2008, 117, 399-410.
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301.
- Boyle, S.; Gilchrist, S.; Bridger, J.M.; Mahy, N.L.; Ellis, J.A.; Bickmore, W.A. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 2001, 10, 211–219.
- Federico, C.; Cantarella, C.D.; Di Mare, P.; Tosi, S.; Saccone, S. The radial arrangement of the human chromosome 7 in the lymphocyte cell nucleus is associated with chromosomal band gene density. Chromosoma 2008, 117, 399–410.
- Croft, J.; Bridger, J.M.; Boyle, S.; Perry, P.; Teague, P.; Bickmore, W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999, 145, 1119–1131.
- Malhas, A.; Lee, C.F.; Sanders, R.; Saunders, N.J.; Vaux, D.J. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 2007, 176, 593–603.
- Tanabe, H.; Mùller, S.; Neusser, M.; von Hase, J.; Calcagno, E.; Cremer, M.; Solovei, S.; Cremer, C.; Cremer, T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA 2002, 99, 4424–4429.
- Saccone, S.; De Sario, A.; Della Valle, G.; Bernardi, G. The highest gene concentrations in the human genome are in telomeric bands of metaphase chromosomes. Proc. Natl. Acad. Sci. USA 1992, 89, 4913–4917.
- Saccone, S.; De Sario, A.; Wiegant, J.; Raap, A.K.; Della Valle, G.; Bernardi, G. Correlations between isochores and chromosomal bands in the human genome. Proc. Natl. Acad. Sci. USA 1993, 90, 11929–11933.
- Saccone, S.; Federico, C.; Solovei, I.; Croquette, M.F.; Della Valle, G.; Bernardi, G. Identification of the gene-richest bands in human prometaphase chromosomes. Chromosome Res. 1999, 7, 379–386.
- Federico, C.; Andreozzi, L.; Saccone, S.; Bernardi, G. Gene density in the Giemsa bands of human chromosomes. Chromosome Res. 2000, 8, 737–746.
- Bickmore, W.A.; van Steensel, B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell 2013, 152, 1270–1284.
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306–1311.
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006, 38, 1348–1354.
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309.
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293.
- Kurz, A.; Lampel, S.; Nickolenko, J.E.; Bradl, J.; Benner, A.; Zirbel, R.M.; Cremer, T.; Lichter, P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 1996, 135, 1195-1205.
- Dietzel, S.; Schiebel, K.; Little, G.; Edelmann, P.; Rappold, G.A.; Eils, R.; Cremer, C.; Cremer, T. The 3D Positioning of ANT2 and ANT3 Genes within Female X Chromosome Territories Correlates with Gene Activity. Exp. Cell Res. 1999, 252, 363-375.
- Scheuermann, M.O.; Tajbakhsh, J.; Kurz, A.; Saracoglu, K.; Eils, R.; Lichter, P. Topology of genes and nontranscribed se-quences in human interphase nuclei. Exp. Cell Res. 2004, 301, 266-279.
- Galiovà, G.; Bàrtovà, E.; Kozubek, S. Nuclear topography of beta-like globin gene cluster in IL-3-stimulated human leu-kemic K-562 cells. Blood Cells Mol. Dis. 2004, 33, 4-14.
- Tajbakhsh, J.; Luz, H.; Bornfleth, H.; Lampel, S.; Cremer, C.; Lichter, P. Spatial Distribution of GC- and AT-Rich DNA Sequences within Human Chromosome Territories. Exp. Cell Res. 2000, 255, 229-237.
- Bickmore, W.A.; van Steensel, B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell 2013, 152, 1270-1284.
- Verschure, P.J.; van der Kraan, I.; Enserink, J.M.; Moné M.J.; Manders, E.M.M.; van Driel, R. Large-scale Chromatin Or-ganization and the Localization of Proteins Involved in Gene Expression in Human Cells. J. Histochem. Cytochem. 2002, 50, 1303-1312.
- Branco, M.R.; Pombo, A. Intermingling of Chromosome Territories in Interphase Suggests Role in Translocations and Transcription-Dependent Associations. PLoS Biol. 2006, 4, e138.
- Volpi, E.V.; Chevret, E.; Jones, T.; Vatcheva, R.; Williamson, J.; Beck, S.; Campbell, R.D.; Goldsworthy, M.; Powis, S.H.; Ragoussis, J.; et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000, 113, 1565-1576.
- Williams, R.R.E.; Broad, S.; Sheer, D.; Ragoussis, J. Subchromosomal Positioning of the Epidermal Differentiation Com-plex (EDC) in Keratinocyte and Lymphoblast Interphase Nuclei. Exp. Cell Res. 2002, 272, 163-175.
- Szczerbal, I.; Foster, H.A.; Bridger, J.M. The spatial repositioning of adipogenesis genes is correlated with their expres-sion status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma 2009, 118, 647-663.
- Khanna, N.; Hu, Y.; Belmont, A.S. HSP70 Transgene Directed Motion to Nuclear Speckles Facilitates Heat Shock Activa-tion. Curr. Biol. 2014, 24, 1138-1144.
- Bourne, G.; Moir, C.; Bikkul, U.; Ahmed Hassan, M.; Kill, I.R.; Eskiw, C.H.; Tosi, S. Interphase chromosome behavior in normal and diseased cells. In: Yurov Y., Vorsanova S., Iourov I. (eds) Human Interphase Chromosomes. Springer, New York, NY. 2013, 9-33.
- Lahbib-Mansais, Y.; Barasc, H.; Marti-Marimon, M.; Mompart, F.; Iannuccelli, E.; Robelin, D.; Riquet, J.; Yerle-Bouissou, M. Expressed alleles of imprinted IGF2, DLK1 and MEG3 colocalize in 3Dpreserved nuclei of porcine fetal cells. BMC Cell. Biol. 2016, 17, 35.
- Harewood, L.; Fraser, P. The impact of chromosomal rearrangements on regulation of gene expression. Hum. Mol. Genet. 2014, 23, R76-R82.
- Gheldof, N.; Witwicki, R.; Migliavacca, E.; Leleu, M.; Didelot, G.; Harewood, L.; Rougemont, J.; Reymond, A. Structural variation-associated expression changes are paralleled by chromatin architecture modifications. PLoS One 2013, 8, e79973.
