Biologics can be re-engineered for blood-brain barrier (BBB) transport as IgG fusion proteins, where the IgG domain is a monoclonal antibody (MAb) that targets an endogenous BBB transporter, such as the insulin receptor (IR) or transferrin receptor (TfR). The IR and TfR at the BBB transport the receptor-specific MAb in parallel with the transport of the endogenous ligand, insulin or transferrin.
- blood–brain barrier
- brain drug delivery
- monoclonal antibody
- transferrin receptor
- insulin receptor
- Structure of the Human Transferrin Receptor-Holo Transferrin Complex
1. Structure of the Human Transferrin Receptor-Holo Transferrin Complex
Transferrin (Tf) is a 679 amino acid bilobular protein comprised of an N-lobe (amino acids 1–331) and a C-lobe (amino acids 339–679), joined by a short linker (amino acids 332–338), and both lobes bind 1 ferrous (Fe+3) atom [37][1]. There are two transferrin receptors, TfR1 and TfR2, which are products of separate genes [38][2]. The TfR expressed at the BBB was identified with a BBB genomics investigation as TfR1 [39][3]. The crystal structure at a resolution of 3.2 angstroms was reported for the complex of the human TfR1 extracellular domain (ECD) and holo-Tf [37][1]. The human TfR1 ECD was expressed in baby hamster kidney (BHK) fibroblasts, and the human Tf was also expressed in BHK cells [37][1]. The Tf was mutated (Y426F, Y517F) to eliminate iron binding to the C-lobe, and the Tf was also mutated (N413D, N611D) to eliminate Tf N-linked glycosylation [37][1]. The hetero-tetrameric Tf-TfR complex is formed by two receptors and two holo-Tf molecules [37][1]. The TfR1 is a 760 amino acid protein comprised of multiple domains, including the intracellular amino terminal domain (amino acids 1–67), the transmembrane domain (amino acids 68–88), a stalk domain, which forms disulfide bonds between two receptors (amino acids 89–120), two protease-like domains (amino acids 121–188 and 384–606), an apical domain (amino acids 189–383), and a helical domain (amino acids 607–760) [37][1]. Amino acids 121–760 form the monomeric ECD of the TfR1. Transferrin in plasma exists in three forms: about 40% is apo-Tf, which does not bind to the TfR1 at physiologic pH; about 30% is diferric holo-Tf; and about 30% is mono-ferric Tf [37][1]. The affinity of diferric Tf for the TfR1 is ~6-fold greater than the affinity of monoferric Tf [40][4]. The concentration of Tf in human plasma is 45,000 nM [41][5], and the concentration of holo-Tf is about 25,000 nM. The plasma concentration of holo-Tf is nearly 1000-fold greater than the TfR1 concentration at the brain capillary endothelium in vivo, which is 40 nM [35][6]. The optimal TfRMAb binding site on the TfR for a monoclonal antibody (MAb) against the TfR is the is the apical domain, as holo-Tf binds to the protease-like and helical domains of the TfR1 as shown in Figure 1A.
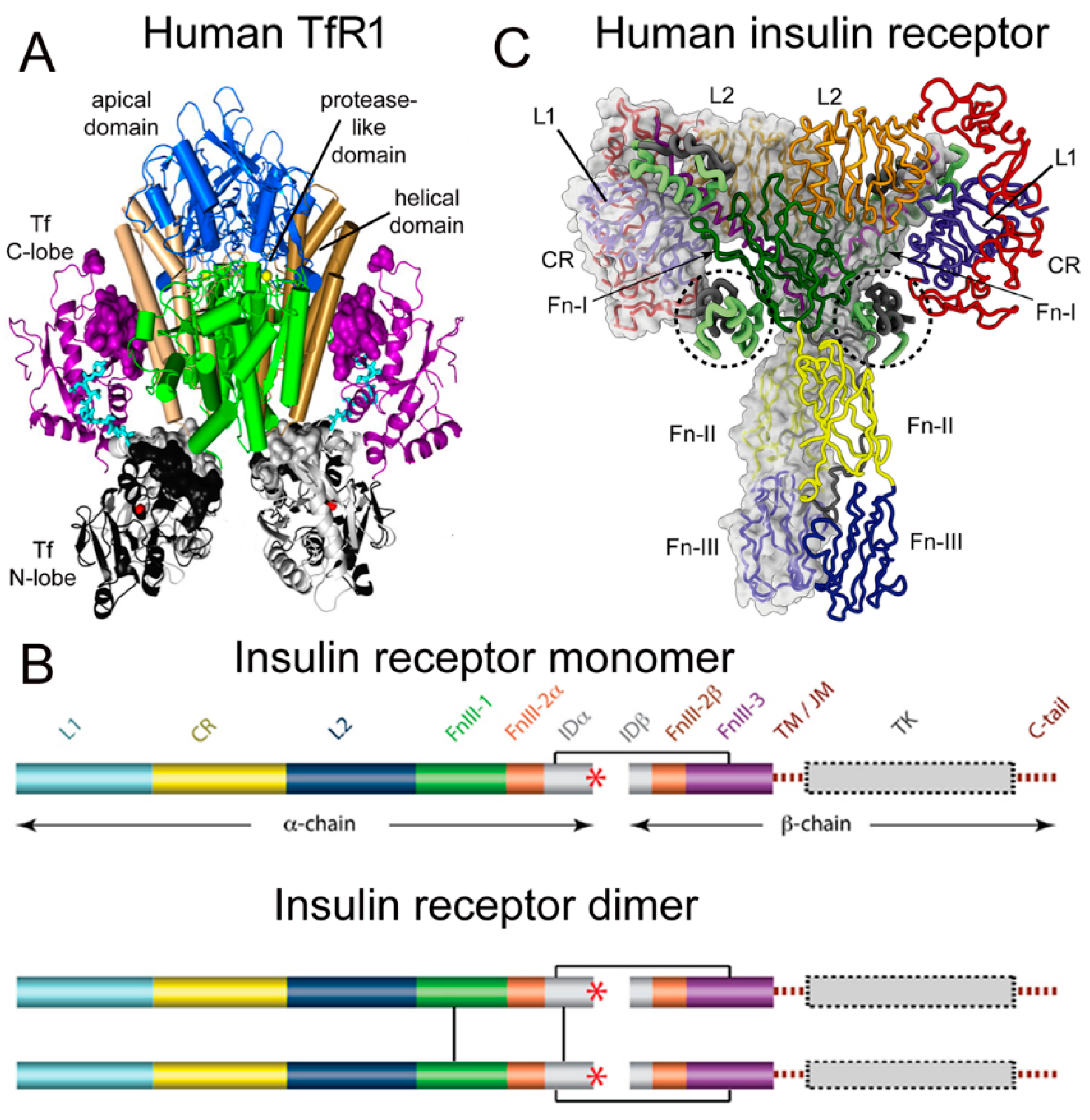 Figure 1. (A) Three-dimensional structure of the complex of the human TfR ECD and holo-Tf. The tetrameric complex is comprised of 2 TfRs and 2 holo-Tf molecules. The cell surface is at the bottom of the structure and the apical domain (blue) is at the top; the 2 protease-like domains are shown in green and the helical domain is shown in brown/tan. The N-lobe and C-lobe of Tf are shown in gray/black and purple, respectively. The Fe+3 bound within the N-lobe is shown in red; the linker between the N and C lobes of Tf is cyan. Reproduced with permission from [37][1]. (B) Two-dimensional structure of the human IR as a monomer (top) and a dimer (bottom). A single disulfide bond joins the alpha and beta chains of each monomer, and the dimer is formed by 2 disulfide bonds between each alpha chain. Reproduced from [42][7], Copyright© 2011 licensed under Creative Commons Attribution License (CC-BY). (C) Three-dimensional structure of the complex of the human IR and insulin. The structure is comprised of the IR dimer and 4 bound insulin molecules. Insulin bound to the second site formed by the FnIII-1/FnIII-2 domains is encircled. Reproduced with permission from [43][8], Copyright© 2021 Elsevier, as reported in [44][9]. The IR domains in panels B and C are defined in the text.
Figure 1. (A) Three-dimensional structure of the complex of the human TfR ECD and holo-Tf. The tetrameric complex is comprised of 2 TfRs and 2 holo-Tf molecules. The cell surface is at the bottom of the structure and the apical domain (blue) is at the top; the 2 protease-like domains are shown in green and the helical domain is shown in brown/tan. The N-lobe and C-lobe of Tf are shown in gray/black and purple, respectively. The Fe+3 bound within the N-lobe is shown in red; the linker between the N and C lobes of Tf is cyan. Reproduced with permission from [37][1]. (B) Two-dimensional structure of the human IR as a monomer (top) and a dimer (bottom). A single disulfide bond joins the alpha and beta chains of each monomer, and the dimer is formed by 2 disulfide bonds between each alpha chain. Reproduced from [42][7], Copyright© 2011 licensed under Creative Commons Attribution License (CC-BY). (C) Three-dimensional structure of the complex of the human IR and insulin. The structure is comprised of the IR dimer and 4 bound insulin molecules. Insulin bound to the second site formed by the FnIII-1/FnIII-2 domains is encircled. Reproduced with permission from [43][8], Copyright© 2021 Elsevier, as reported in [44][9]. The IR domains in panels B and C are defined in the text.
- Structure of the Human Insulin Receptor-Insulin Complex
2. Structure of the Human Insulin Receptor-Insulin Complex
There are two human insulin receptors, designated IR-A (short form) and IR-B (long form), which are derived from a single gene by alternate processing of the primary transcript. In IR-A, which is primarily expressed in cancer and fetal tissues [45][10], exon 11 is deleted, resulting in a 12 amino acid truncation at the carboxyl terminus of the alpha chain, which corresponds to the α-CT domain of IR-B. IR-B is the isoform predominantly expressed in tissues [45][10]. Following removal of a 27 amino acid signal peptide, IR-B is encoded as a 1355 amino acid polypeptide, which is proteolytically cleaved to the alpha chain, amino acids 1–731 (not counting the signal peptide), and the beta chain, amino acids 736–1355 [46][11]. This separation into alpha and beta chains occurs at a furin cleavage site, RKRR [47][12], which corresponds to amino acids 732–735, and this sequence is removed in the cleavage. The cleavage into the separate alpha and beta chains is shown in Figure 1B (top). The alpha chain is formed by the first leucine-rich (L1) domain, the cysteine-rich (CR) domain, the second leucine-rich (L2) domain, the first fibronection (Fn) III domain (FnIII-1), the first part of the second fibronection III domain (FnIII-2α), and the first part of the insert domain (IDα); the final 12 amino acids of the alpha chain is the αCT domain, which is involved in insulin binding [46][11]. The beta chain is formed by the second part of the insert domain (IDβ), the second part of the FnIII-2 domain (FnIII-2β), the third fibronectin domain (FnIII-3), the transmembrane (TM) domain, the juxtamembrane (JM) domain, the tyrosine kinase (TK) domain, and the carboxyl terminus (Figure 1B, top). An inter-chain disulfide bond joins the alpha and beta chains, and two additional disulfides between the two alpha chains form the hetero-tetrameric structure of the IR (Figure 1B, bottom). The ECD of the IR, which is approximately 900 amino acids in length, is formed by cleavage near the TM domain and includes all of the alpha chain and the amino terminal portion of the beta chain. The crystal structure of the ECD of the human IR complexed with monoclonal antibodies was originally produced [48][13]. Recently, the three-dimensional structure of the complex of insulin and the IR tetrameric structure was generated with cryo electron microscopy [44[9][14],49], as recently reviewed [43][8], and this structure is shown in Figure 1C. The structure of the insulin/IR complex reveals each IR monomer binds two insulin molecules, so that the IR dimer shown in Figure 1C binds four insulin molecules; two insulins are bound to the classical high-affinity binding site formed by interaction of the L1 and αCT domains of each alpha subunit and two insulins are bound to a low-affinity second site formed by interactions of the FnIII-1 and FnIII-2 domains of each alpha subunit (Figure 1C). Insulin is synthesized as a proinsulin precursor in pancreatic beta cells, and proinsulin is cleaved to 2 insulin subunits, the 21 amino acid A-chain and the 30 amino acid B-chain, which are joined together by 2 disulfide bonds [43][8]. The fasting plasma insulin concentration is about 0.3 nM in humans and primates [50,51][15][16]. The plasma concentration of insulin is ~100-fold lower than the IR concentration at the brain capillary endothelium in vivo, which is 24 nM [35][6].
- BBB Transport of Holo-Transferrin
3. BBB Transport of Holo-Transferrin
The model solutions by numerical analysis of a partly flow-partly compartmental model of BBB holo-Tf transport have been described previously [35][6], and the holo-Tf model is shown in Figure 2.
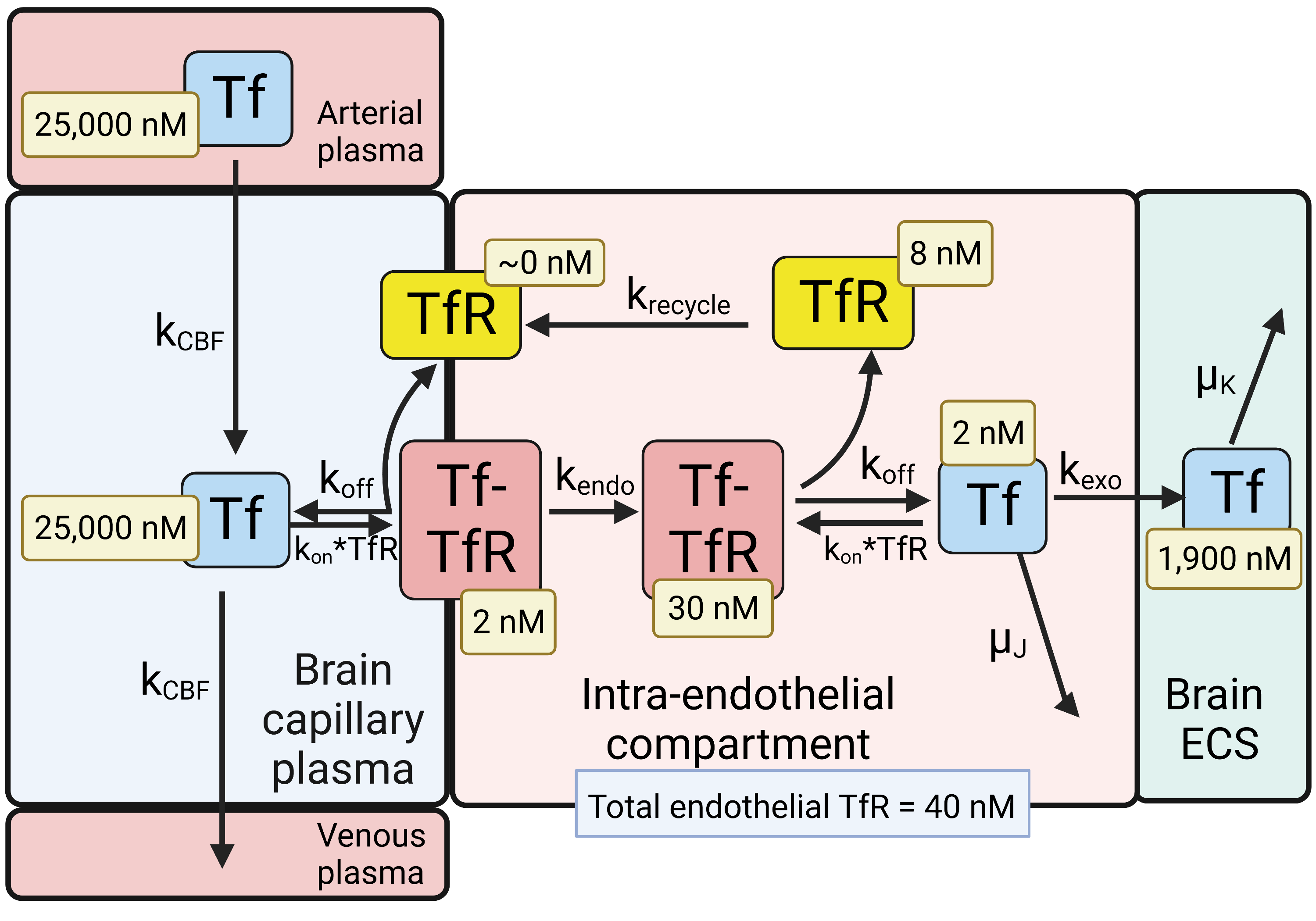
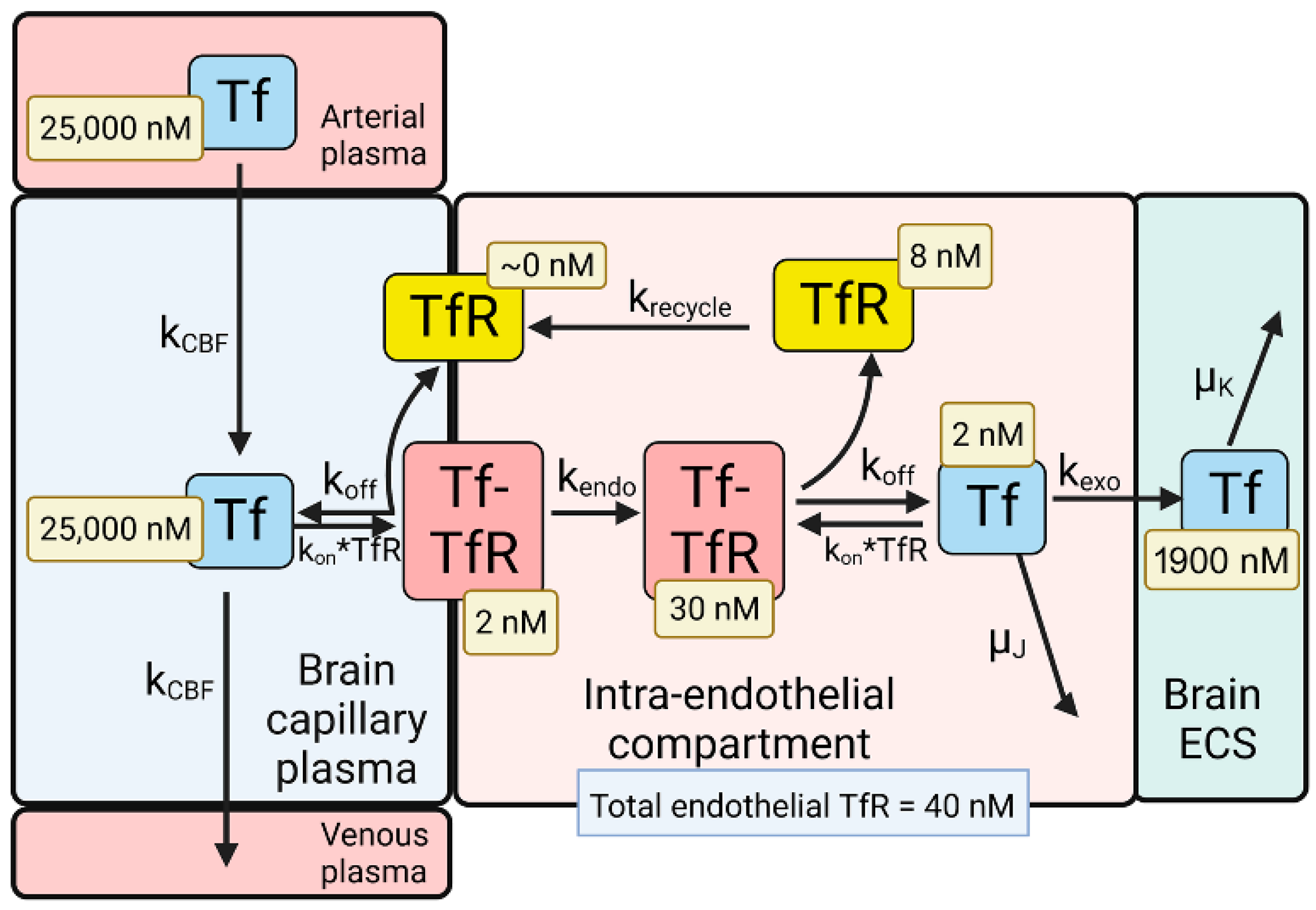
Figure 2. Model of transport of holo-transferrin (Tf) from the blood to the brain extracellular space (ECS) through the brain capillary endothelium, which forms the BBB in vivo. Holo-Tf in plasma binds the transferrin receptor (TfR) on the luminal endothelial membrane to form the luminal Tf-TfR complex, which is followed by endocytosis into the intra-endothelial compartment. Following dissociation of the Tf within the endothelium, the Tf undergoes exocytosis into the brain extracellular space (ECS). The model allows for estimations of the concentrations of Tf, or the TfR, in each pool in the transcytosis pathway, and these concentrations are shown in the light-yellow boxes. Adapted from [35], Copyright© 2021 licensed under Creative Commons Attribution License (CC-BY). Image created with Biorender.com.
The dissociation (k
off
on
−1
−1
−1
D = 0.6 nM [52]. The rate constants of endocytosis (k
endo
exo
recycle
CBF
−1
1/2
−1
1/2
−1
1/2
−1 [35]. The initial conditions of the model set [Tf] = 25,000 nM and [Tf] = 0 for the concentration of Tf in the plasma and brain ECS, respectively. The experimentally observed concentration of Tf in the brain is 114 ug/gram [53], which is equal to 2000 nM, as the brain water volume is 0.7 mL/g [54]. Given a rate constant of Tf degradation in the brain of μ
K
−1
1/2
1/2
1/2 of Tf, which is 2.5 days [55]. At steady state, the concentration of free TfR on the luminal membrane was ~0 (
Figure 2), owing to the vastly greater concentration of holo-Tf in plasma, 25,000 nM, as compared to the total concentration of TfR, 40 nM, at the brain capillary endothelium [35]. Most of the endothelial TfR, 30 nM or 75% of total endothelial TfR, was localized to the intra-endothelial compartment as a Tf-TfR complex; the concentration of free Tf and free TfR within the endothelial compartment was estimated to be 2 and 8 nM, respectively (
Figure 2
Figure 2
Figure 1A), which is embedded in the endothelial plasma membrane.
- BBB Transport of Insulin
A partly flow-partly compartmental model of BBB transport of insulin is outlined in
4. BBB Transport of Insulin
Figure 3.
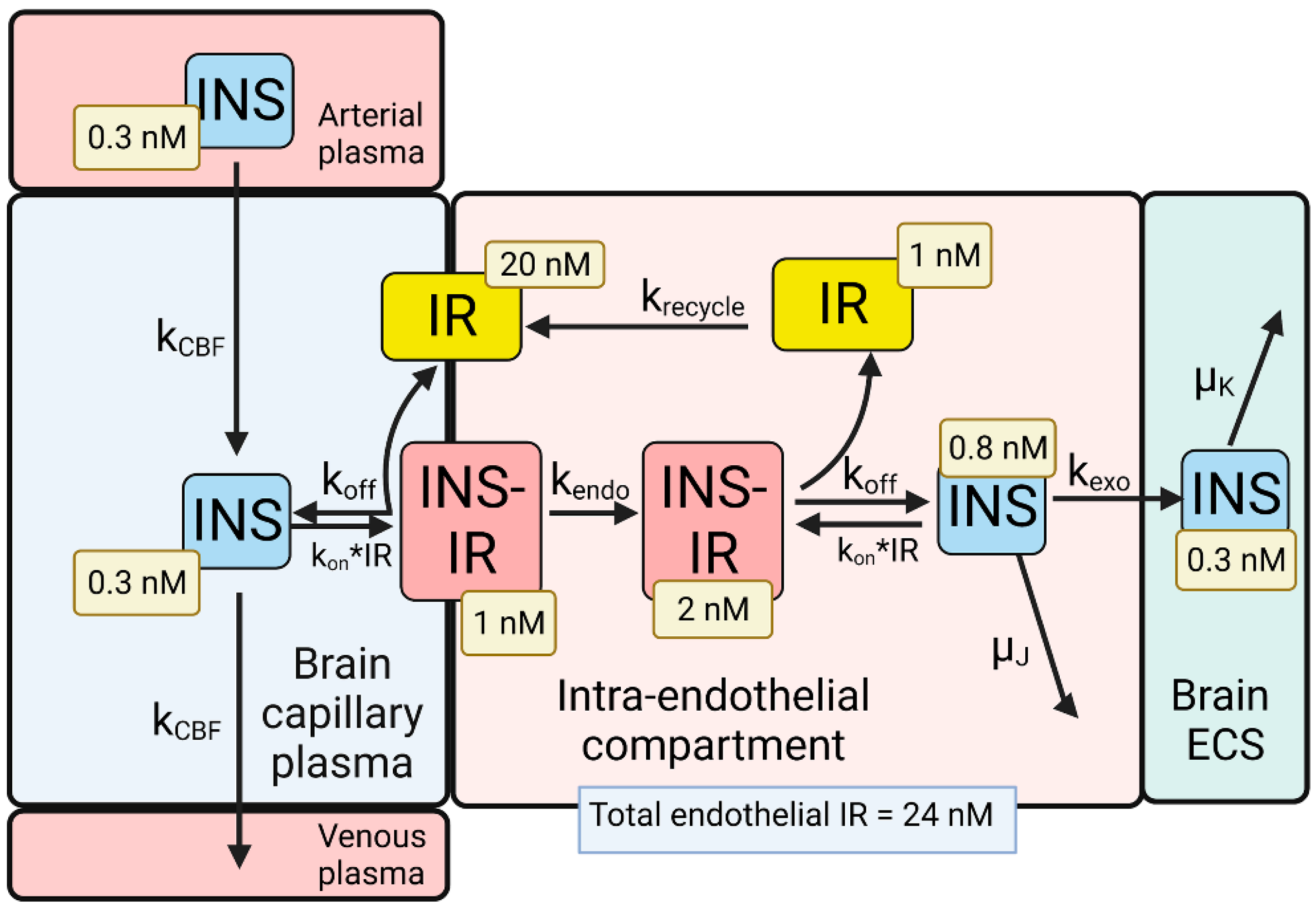
Figure 3. Model of transport of insulin (INS) from the blood to the brain extracellular space (ECS) through the brain capillary endothelium, which forms the BBB in vivo. INS in plasma binds the insulin receptor (IR) on the luminal endothelial membrane to form the luminal INS-IR complex, which is followed by endocytosis into the intra-endothelial compartment. Following dissociation of the INS within the endothelium, the INS undergoes exocytosis into the brain ECS. The model allowed for estimations of the concentrations of INS, or the IR, in each pool in the transcytosis pathway, and these concentrations are shown in the light-yellow boxes. Adapted from [35], Copyright© 2021 licensed under Creative Commons Attribution License (CC-BY). Image created with Biorender.com.
The differential equations and insulin (INS) model solutions by numerical analysis have been described previously [35]. The dissociation (k
off
on
−1
−1
−1
D = 2.6 nM [56]. The rate constants of endocytosis (k
endo
exo
recycle
−1
1/2
−1
1/2
−1
1/2 = 20 min), respectively, as described previously [35]. The rate constant of cerebral blood flow (k
CBF
−1
1/2 = 1 s), was derived from the Vp/CBF ratio, where the brain plasma volume (Vp) is 0.01 mL/g [57], and the rate of cerebral blood flow (CBF) is 0.6 mL/min/g [58]. The rate constant of INS degradation within the endothelium, μ
J
−1
1/2 = 2 h), as prior work showed no insulin degradation by isolated human brain microvessels within 60 min at 37 °C [39]. The initial conditions of the model set [INS] = 0.3 nM and [INS] = 0 for the concentration of INS in the plasma and brain ECS, respectively. If the model was run from 0 to 6 h, and the rate constant of INS degradation in brain was set at μ
K
−1
1/2
Figure 3), which corresponds to the experimentally observed insulin concentration in the brain. The brain insulin concentration is 9.6 ± 3.4 μU/g [59], which is equivalent to 48 μU/mL, given an ECS volume in the brain of 0.2 mL/g [60]. Converting μU of insulin to fmol of insulin, based on 1 μU = 6 fmol [61], the experimentally observed brain insulin concentration is 0.3 nM. A T
1/2
1/2 of INS removal from plasma, which is 4–6 min [62]. At steady state, the concentration of free IR on the luminal membrane was 20 nM (
Figure 3), which is 83% of the total IR, 24 nM, in the brain capillary endothelium [35]. The high concentration of free IR at the luminal membrane (
Figure 3
Figure 2
Figure 3). These modeling studies for INS indicate an IRMAb in the plasma primarily binds to the unbound IR, rather than the INS-IR complex, where IRMAb is a MAb against the IR.
- References
[35] Pardridge, W.M.; Chou, T. Mathematical Models of Blood-Brain Barrier Transport of Monoclonal Antibodies Targeting the Transferrin Receptor and the Insulin Receptor. Pharmaceuticals (Basel) 2021, 14, doi:10.3390/ph14060535.
[36] Boado, R.J.; Pardridge, W.M. Brain and Organ Uptake in the Rhesus Monkey in Vivo of Recombinant Iduronidase Compared to an Insulin Receptor Antibody-Iduronidase Fusion Protein. Mol Pharm 2017, 14, 1271-1277, doi:10.1021/acs.molpharmaceut.6b01166
[37] Eckenroth, B.E.; Steere, A.N.; Chasteen, N.D.; Everse, S.J.; Mason, A.B. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc Natl Acad Sci U S A 2011, 108, 13089-13094, doi:10.1073/pnas.1105786108.
[38] Herbison, C.E.; Thorstensen, K.; Chua, A.C.; Graham, R.M.; Leedman, P.; Olynyk, J.K.; Trinder, D. The role of transferrin receptor 1 and 2 in transferrin-bound iron uptake in human hepatoma cells. Am J Physiol Cell Physiol 2009, 297, C1567-1575, doi:10.1152/ajpcell.00649.2008.
[39] Pardridge, W.M. The Isolated Brain Microvessel: A Versatile Experimental Model of the Blood-Brain Barrier. Front Physiol 2020, 11, 398, doi:10.3389/fphys.2020.00398.
[40] Mason, A.B.; Byrne, S.L.; Everse, S.J.; Roberts, S.E.; Chasteen, N.D.; Smith, V.C.; MacGillivray, R.T.; Kandemir, B.; Bou-Abdallah, F. A loop in the N-lobe of human serum transferrin is critical for binding to the transferrin receptor as revealed by mutagenesis, isothermal titration calorimetry, and epitope mapping. J Mol Recognit 2009, 22, 521-529, doi:10.1002/jmr.979.
[41] Schmaier, A.H. Transferrin: a blood coagulation modifier. Cell Res 2020, 30, 101-102, doi:10.1038/s41422-020-0275-z.
[42] Ward, C.W.; Lawrence, M.C. Landmarks in insulin research. Front Endocrinol (Lausanne) 2011, 2, 76, doi:10.3389/fendo.2011.00076.
[43] Lawrence, M.C. Understanding insulin and its receptor from their three-dimensional structures. Mol Metab 2021, 52, 101255, doi:10.1016/j.molmet.2021.101255.
[44] Gutmann, T.; Schafer, I.B.; Poojari, C.; Brankatschk, B.; Vattulainen, I.; Strauss, M.; Coskun, U. Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J Cell Biol 2020, 219, doi:10.1083/jcb.201907210.
[45] Giudice, J.; Barcos, L.S.; Guaimas, F.F.; Penas-Steinhardt, A.; Giordano, L.; Jares-Erijman, E.A.; Coluccio Leskow, F. Insulin and insulin like growth factor II endocytosis and signaling via insulin receptor B. Cell Commun Signal 2013, 11, 18, doi:10.1186/1478-811X-11-18.
[46] Menting, J.G.; Whittaker, J.; Margetts, M.B.; Whittaker, L.J.; Kong, G.K.; Smith, B.J.; Watson, C.J.; Zakova, L.; Kletvikova, E.; Jiracek, J., et al. How insulin engages its primary binding site on the insulin receptor. Nature 2013, 493, 241-245, doi:10.1038/nature11781.
[47] Bravo, D.A.; Gleason, J.B.; Sanchez, R.I.; Roth, R.A.; Fuller, R.S. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J Biol Chem 1994, 269, 25830-25837.
[48] McKern, N.M.; Lawrence, M.C.; Streltsov, V.A.; Lou, M.Z.; Adams, T.E.; Lovrecz, G.O.; Elleman, T.C.; Richards, K.M.; Bentley, J.D.; Pilling, P.A., et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 2006, 443, 218-221, doi:10.1038/nature05106.
[49] Uchikawa, E.; Choi, E.; Shang, G.; Yu, H.; Bai, X.C. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife 2019, 8, doi:10.7554/eLife.48630.
[50] Bar, R.S.; Gorden, P.; Roth, J.; Kahn, C.R.; De Meyts, P. Fluctuations in the affinity and concentration of insulin receptors on circulating monocytes of obese patients: effects of starvation, refeeding, and dieting. J Clin Invest 1976, 58, 1123-1135, doi:10.1172/JCI108565.
[51] Bremer, A.A.; Stanhope, K.L.; Graham, J.L.; Cummings, B.P.; Wang, W.; Saville, B.R.; Havel, P.J. Fructose-fed rhesus monkeys: a nonhuman primate model of insulin resistance, metabolic syndrome, and type 2 diabetes. Clin Transl Sci 2011, 4, 243-252, doi:10.1111/j.1752-8062.2011.00298.x.
[52] Giannetti, A.M.; Bjorkman, P.J. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J Biol Chem 2004, 279, 25866-25875, doi:10.1074/jbc.M401467200.
[53] Dodd, P.R.; Eckert, A.L.; Fletcher, L.M.; Kril, J.J.; Harper, C.G.; Halliday, J.W. Concentrations of transferrin and carbohydrate-deficient transferrin in postmortem human brain from alcoholics. Addict Biol 1997, 2, 337-348, doi:10.1080/13556219772633.
[54] Sigurdsson, S.; Aspelund, T.; Forsberg, L.; Fredriksson, J.; Kjartansson, O.; Oskarsdottir, B.; Jonsson, P.V.; Eiriksdottir, G.; Harris, T.B.; Zijdenbos, A., et al. Brain tissue volumes in the general population of the elderly: the AGES-Reykjavik study. Neuroimage 2012, 59, 3862-3870, doi:10.1016/j.neuroimage.2011.11.024.
[55] Strahan, M.E.; Crowe, A.; Morgan, E.H. Iron uptake in relation to transferrin degradation in brain and other tissues of rats. Am J Physiol 1992, 263, R924-929, doi:10.1152/ajpregu.1992.263.4.R924.
[56] Scapin, G.; Dandey, V.P.; Zhang, Z.; Prosise, W.; Hruza, A.; Kelly, T.; Mayhood, T.; Strickland, C.; Potter, C.S.; Carragher, B. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 2018, 556, 122-125, doi:10.1038/nature26153.
[57] Mandikian, D.; Figueroa, I.; Oldendorp, A.; Rafidi, H.; Ulufatu, S.; Schweiger, M.G.; Couch, J.A.; Dybdal, N.; Joseph, S.B.; Prabhu, S., et al. Tissue Physiology of Cynomolgus Monkeys: Cross-Species Comparison and Implications for Translational Pharmacology. AAPS J 2018, 20, 107, doi:10.1208/s12248-018-0264-z.
[58] Joris, P.J.; Mensink, R.P.; Adam, T.C.; Liu, T.T. Cerebral Blood Flow Measurements in Adults: A Review on the Effects of Dietary Factors and Exercise. Nutrients 2018, 10, doi:10.3390/nu10050530.
[59] Frank, H.J.; Jankovic-Vokes, T.; Pardridge, W.M.; Morris, W.L. Enhanced insulin binding to blood-brain barrier in vivo and to brain microvessels in vitro in newborn rabbits. Diabetes 1985, 34, 728-733, doi:10.2337/diab.34.8.728.
[60] Sykova, E.; Nicholson, C. Diffusion in brain extracellular space. Physiol Rev 2008, 88, 1277-1340, doi:10.1152/physrev.00027.2007.
[61] Knopp, J.L.; Holder-Pearson, L.; Chase, J.G. Insulin Units and Conversion Factors: A Story of Truth, Boots, and Faster Half-Truths. J Diabetes Sci Technol 2019, 13, 597-600, doi:10.1177/1932296818805074.
[62] Duckworth, W.C.; Bennett, R.G.; Hamel, F.G. Insulin degradation: progress and potential. Endocr Rev 1998, 19, 608-624, doi:10.1210/edrv.19.5.0349.
References
- Eckenroth, B.E.; Steere, A.N.; Chasteen, N.D.; Everse, S.J.; Mason, A.B. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc. Natl. Acad. Sci. USA 2011, 108, 13089–13094.
- Herbison, C.E.; Thorstensen, K.; Chua, A.C.; Graham, R.M.; Leedman, P.; Olynyk, J.K.; Trinder, D. The role of transferrin receptor 1 and 2 in transferrin-bound iron uptake in human hepatoma cells. Am. J. Physiol. Cell. Physiol. 2009, 297, C1567–C1575.
- Pardridge, W.M. The Isolated Brain Microvessel: A Versatile Experimental Model of the Blood-Brain Barrier. Front. Physiol. 2020, 11, 398.
- Mason, A.B.; Byrne, S.L.; Everse, S.J.; Roberts, S.E.; Chasteen, N.D.; Smith, V.C.; MacGillivray, R.T.; Kandemir, B.; Bou-Abdallah, F. A loop in the N-lobe of human serum transferrin is critical for binding to the transferrin receptor as revealed by mutagenesis, isothermal titration calorimetry, and epitope mapping. J. Mol. Recognit. 2009, 22, 521–529.
- Schmaier, A.H. Transferrin: A blood coagulation modifier. Cell. Res. 2020, 30, 101–102.
- Pardridge, W.M.; Chou, T. Mathematical Models of Blood-Brain Barrier Transport of Monoclonal Antibodies Targeting the Transferrin Receptor and the Insulin Receptor. Pharmaceuticals 2021, 14, 535.
- Ward, C.W.; Lawrence, M.C. Landmarks in insulin research. Front. Endocrinol. (Lausanne) 2011, 2, 76.
- Lawrence, M.C. Understanding insulin and its receptor from their three-dimensional structures. Mol. Metab. 2021, 52, 101255.
- Gutmann, T.; Schafer, I.B.; Poojari, C.; Brankatschk, B.; Vattulainen, I.; Strauss, M.; Coskun, U. Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J. Cell. Biol. 2020, 219, e201907210.
- Giudice, J.; Barcos, L.S.; Guaimas, F.F.; Penas-Steinhardt, A.; Giordano, L.; Jares-Erijman, E.A.; Coluccio Leskow, F. Insulin and insulin like growth factor II endocytosis and signaling via insulin receptor B. Cell. Commun. Signal. 2013, 11, 18.
- Menting, J.G.; Whittaker, J.; Margetts, M.B.; Whittaker, L.J.; Kong, G.K.; Smith, B.J.; Watson, C.J.; Zakova, L.; Kletvikova, E.; Jiracek, J.; et al. How insulin engages its primary binding site on the insulin receptor. Nature 2013, 493, 241–245.
- Bravo, D.A.; Gleason, J.B.; Sanchez, R.I.; Roth, R.A.; Fuller, R.S. Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J. Biol. Chem. 1994, 269, 25830–25837.
- McKern, N.M.; Lawrence, M.C.; Streltsov, V.A.; Lou, M.Z.; Adams, T.E.; Lovrecz, G.O.; Elleman, T.C.; Richards, K.M.; Bentley, J.D.; Pilling, P.A.; et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 2006, 443, 218–221.
- Uchikawa, E.; Choi, E.; Shang, G.; Yu, H.; Bai, X.C. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife 2019, 8, e48630.
- Bar, R.S.; Gorden, P.; Roth, J.; Kahn, C.R.; De Meyts, P. Fluctuations in the affinity and concentration of insulin receptors on circulating monocytes of obese patients: Effects of starvation, refeeding, and dieting. J. Clin. Investig. 1976, 58, 1123–1135.
- Bremer, A.A.; Stanhope, K.L.; Graham, J.L.; Cummings, B.P.; Wang, W.; Saville, B.R.; Havel, P.J. Fructose-fed rhesus monkeys: A nonhuman primate model of insulin resistance, metabolic syndrome, and type 2 diabetes. Clin. Transl. Sci. 2011, 4, 243–252.
- Giannetti, A.M.; Bjorkman, P.J. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J. Biol. Chem. 2004, 279, 25866–25875.
- Dodd, P.R.; Eckert, A.L.; Fletcher, L.M.; Kril, J.J.; Harper, C.G.; Halliday, J.W. Concentrations of transferrin and carbohydrate-deficient transferrin in postmortem human brain from alcoholics. Addict. Biol. 1997, 2, 337–348.
- Sigurdsson, S.; Aspelund, T.; Forsberg, L.; Fredriksson, J.; Kjartansson, O.; Oskarsdottir, B.; Jonsson, P.V.; Eiriksdottir, G.; Harris, T.B.; Zijdenbos, A.; et al. Brain tissue volumes in the general population of the elderly: The AGES-Reykjavik study. Neuroimage 2012, 59, 3862–3870.
- Strahan, M.E.; Crowe, A.; Morgan, E.H. Iron uptake in relation to transferrin degradation in brain and other tissues of rats. Am. J. Physiol. 1992, 263, R924–R929.
- Scapin, G.; Dandey, V.P.; Zhang, Z.; Prosise, W.; Hruza, A.; Kelly, T.; Mayhood, T.; Strickland, C.; Potter, C.S.; Carragher, B. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 2018, 556, 122–125.
- Mandikian, D.; Figueroa, I.; Oldendorp, A.; Rafidi, H.; Ulufatu, S.; Schweiger, M.G.; Couch, J.A.; Dybdal, N.; Joseph, S.B.; Prabhu, S.; et al. Tissue Physiology of Cynomolgus Monkeys: Cross-Species Comparison and Implications for Translational Pharmacology. AAPS J. 2018, 20, 107.
- Joris, P.J.; Mensink, R.P.; Adam, T.C.; Liu, T.T. Cerebral Blood Flow Measurements in Adults: A Review on the Effects of Dietary Factors and Exercise. Nutrients 2018, 10, 530.
- Frank, H.J.; Jankovic-Vokes, T.; Pardridge, W.M.; Morris, W.L. Enhanced insulin binding to blood-brain barrier in vivo and to brain microvessels in vitro in newborn rabbits. Diabetes 1985, 34, 728–733.
- Sykova, E.; Nicholson, C. Diffusion in brain extracellular space. Physiol. Rev. 2008, 88, 1277–1340.
- Knopp, J.L.; Holder-Pearson, L.; Chase, J.G. Insulin Units and Conversion Factors: A Story of Truth, Boots, and Faster Half-Truths. J. Diabetes Sci. Technol. 2019, 13, 597–600.
- Duckworth, W.C.; Bennett, R.G.; Hamel, F.G. Insulin degradation: Progress and potential. Endocr. Rev. 1998, 19, 608–624.
