You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Hua Zhang.
LCD1 mutation caused earlier leaf senescence, whereas LCD1 overexpression significantly delayed leaf senescence compared with the wild type in 10-week tomato seedlings. Moreover, LCD1 overexpression was found to delay dark-induced senescence in detached tomato leaves, and the lcd1 mutant showed accelerated senescence.
- tomato
- hydrogen sulfide
- cysteine desulfhydrase
- leaf senescence
1. Introduction
Leaf senescence represents the final stage of leaf development, which is a genetically controlled process [1]. As leaves age, the decomposition of chloroplast is initiated, accompanied by the catabolism of macromolecules including nucleic acids, proteins, and lipids. The decomposed nutrients then transfer to other developing organs, such as young leaves and growing fruit [2]. Chloroplasts constitute approximately 70% of the total proteins in green leaves and chlorophyll degradation causes the first visible signs of leaf senescence [3]. Thus, the coordinated degradation of chlorophyll is crucial for the breakdown of chloroplasts. Terrestrial plants utilize two types of chlorophyll species (i.e., chlorophyll a and chlorophyll b) for photosynthesis [4]. Chlorophyll b has to be converted to chlorophyll a before it can be processed into the degradation pathway and NON-YELLOW COLORING 1 (NYC1) catalyzes the reduction of chlorophyll b to 7-hydroxymethyl chlorophyll a [5]. Chlorophyll a is further decomposed to pheophytin a by Mg-dechelatase NON-YELLOWINGs/STAY-GREENs (NYEs/SGRs) [6]. Pheophytin a is then hydrolyzed by the pheophytinase PPH to generate pheophorbide a which is further catalyzed by oxygenase PAO to produce a red chlorophyll catabolite (RCC) [7]. Moreover, hundreds of senescence-associated genes (SAGs), whose transcripts increase as leaves age [8[8][9],9], are also involved in the regulation of leaf senescence.
Plant hormones are major players influencing each stage of leaf senescence. For instance, ethylene, abscisic acid (ABA), and so on promote leaf senescence, while cytokinins (CKs), gibberellic acid (GA), and so on delay leaf senescence [10,11,12][10][11][12]. Furthermore, leaf senescence-linked events are often associated with the pronounced accumulation of reactive oxygen species (ROS) [13]. Among them, H2O2 is a well-defined inducer of leaf senescence. Recently, it was reported that transcription factor NAC075 delays leaf senescence by deterring ROS accumulation through directly binding the promoter of the antioxidant enzyme gene catalase 2 (CAT) in Arabidopsis [14]. Hydrogen sulfide (H2S) is an important gasotransmitter in both animals and plants [15]. H2S has not only been implicated in seed germination and root development, but can also enhance plant tolerance to various stresses such as heavy metals, drought, salinity, and cold by enhancing the antioxidant system [16,17,18][16][17][18]. In addition, H2S can extend the shelf life of bananas, grapes, strawberries, tomatoes, and so on [19,20,21,22][19][20][21][22]. The underlining mechanism of H2S in alleviating postharvest senescence may involve the activation of the antioxidant system, the inhibition of ethylene synthesis and the signaling pathway, etc.
H2S is endogenously produced in a precise and regulated manner. Cysteine degradation by cysteine desulfhydrases (CDes) to the formation of sulfide, ammonia, and pyruvate was believed to be an important source of H2S [23]. Plant cells contain different CDes localized in the cytoplasm, plastids, and mitochondria [23]. DES1, an O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity, regulates cysteine homeostasis in Arabidopsis [24]. Recently it was reported that the des1 mutant was more sensitive to drought stress and displayed accelerated leaf senescence, while the leaves of OE-DES1 contained adequate chlorophyll levels accompanied by significantly increased drought resistance, suggesting the role of DES1 in regulating leaf senescence [25].
2. Role of LCD1 in Regulating Tomato Leaf Senescence
To elucidate the possible involvement of LCD1 in regulating leaf senescence, two previously reported tomato lines, lcd1-7 and lcd1-9, with mutations near the PAM sequence were used as lcd1 mutants, and two lines (hereafter called LCD1-oe and LCD1-oe1) with an increased expression of LCD1 under the control of the CaMV 35S promoter were also used. The overexpression efficacy of five LCD1 overexpression lines was verified by RT-qPCR as shown in Figure S1 and the lines LCD1-oe and LCD1-oe1, which showed a higher LCD1 expression, were used in the present study. To confirm the role of LCD1 in catalyzing H2S production, the H2S producing rates were determined in leaves of lcd1 and LCD1-oe lines. The data in Figure 1B suggest that lcd1 leaves had a lower H2S producing rate compared with the wild type, while LCD1 overexpression induced a significantly higher level of the H2S producing rate. Besides, H2S production was also evaluated by lead acetate H2S detection strips, and the results showed that lcd1 leaves produced less H2S and LCD1 overexpression produced more H2S (Figure 1C). After 10 weeks of growth, the LCD1 mutation caused earlier leaf senescence compared with the wild type. In contrast, LCD1 overexpression significantly delayed leaf senescence (Figure 1D).
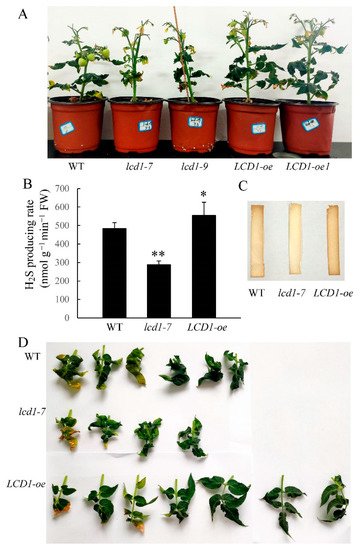

Figure 1. Phenotypic characterization of lcd1 mutants and LCD-oe (over-expression) tomatoes. (A) Phenotype of 10-week-old wild-type (WT), lcd1-7, lcd1-9, LCD1-oe, and LCD1-oe1 plants. (B) H2S producing rate in the mature leaves from wild type, lcd1, and LCD1-oe lines of 10 weeks growth. (C) H2S production from the mature leaves of 10-week-old wild type, lcd1, and LCD1-oe lines detected by lead acetate H2S detection strips (Sigma-Aldrich). (D) The leaves of different tomato lines in (A) were detached and photographed. Data are means of three biological replicates ± standard deviation (SD). The symbols * and ** stand for p < 0.05 and p < 0.01 as determined by the Student’s t-test, respectively.
3. LCD1 Participates in Dark-Induced Senescence
Leaf senescence is an important phenomenon in the growth and development of plant leaves, and darkness is widely used as a tool to induce senescence in detached leaves. To study the role of LCD1 in dark-induced senescence, the mature leaves without visible senescence of 6-week-old wild type, lcd1 mutant, and LCD1-oe were stored in darkness for 8 days. As shown in Figure 2A, lcd1 showed the obvious syndrome of the leaf yellowing phenotype after 5 and 8 days in dark stress, whereas LCD1 overexpression still maintained the green phenotype. To study the kinetics of tomato leaf H2S production during senescence, H2S production in the leaves at different developmental stages—young leaves (YL), mature leaves (ML), senescent leaves (SL), and late senescent leaves (LS)—was evaluated and the H2S detection strips showed browning with senescence, suggesting H2S production increased during leaf senescence (Figure S2). Moreover, H2S production in leaves of wild-type (WT), lcd1, and LCD1-oe tomatoes were also determined during dark-induced senescence (Figure 2B). Generally, an increasing trend of H2S production was observed in all samples during storage, while LCD1-oe leaves showed a higher H2S production compared with the wild-type control. In addition, the lcd1 mutant produced a significantly lower level of H2S compared with the wild type. Therefore, it can be concluded that LCD1 deletion caused a lower H2S release and the attenuated H2S release may cause an accelerated senescence in the lcd1 mutant. Overall, the present results indicate that LCD1 plays a negative role in leaf senescence in both developmental and dark-induced senescence.
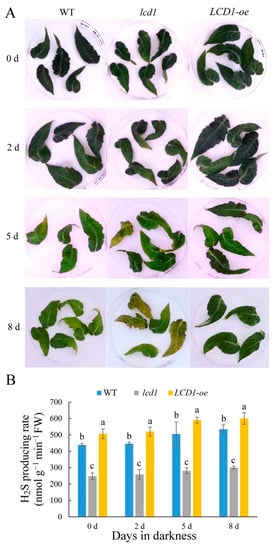

Figure 2. (A) Dark-induced senescence symptoms in detached leaves of 6-week-old wild-type (WT), lcd1, and LCD1-oe tomatoes for up to 8 days. (B) H2S producing rate in detached leaves of 6-week-old wild-type (WT), lcd1, and LCD1-oe tomatoes stored in darkness for up to 8 days. Different letters above the columns stand for significant difference between two values (p < 0.05) at the same time point.
4. Effect of LCD1 on Dark-Triggered Chlorophyll Degradation and Reactive Oxygen Species Accumulation in Detached Tomato Leaves
Chlorophyll degradation is the one of the most significant changes during leaf senescence; thus, chlorophyll contents were determined in wild-type, lcd1 mutant, and LCD1-oe leaves during dark-induced senescence. As shown in Figure 3A, the content of total chlorophyll in the wild type decreased gradually during storage in darkness for 8 days, whereas the content of chlorophyll in the lcd1 mutant showed an obvious decrease on days 5 and 8 under darkness, and the value on day 8 was about 32.6% of the initial value on day 0. In contrast, LCD1 overexpression maintained a relatively higher chlorophyll content compared with the wild type and the lcd1 mutant on days 5 and 8 under darkness. After 8 days in darkness, the chlorophyll content in LCD1 overexpression decreased to 84.6% of the initial value, suggesting the role of LCD1 in delaying dark-induced senescence. As shown in Figure 3B, there were minor changes in the chlorophyll a content between different groups during storage. Moreover, only a slight decrease in chlorophyll a was observed during dark-induced senescence, except for a significant decline found in lcd1 on day 8. Figure 3C shows the change pattern of chlorophyll b content in wild type, lcd1 mutant, and LCD1-oe during dark-induced senescence. With the increase of storage days, the chlorophyll b content decreased in each group. At day 0, chlorophyll b content in lcd1 leaves was about 53.4% of that in the LCD1-oe group, and decreased to 21.3% on day 8 compared with the value on day 0. Furthermore, the ratio of chlorophyll a/b was also evaluated in dark-stored detached leaves of wild-type, lcd1, and LCD1-oe tomatoes for 0, 2, 5, and 8 days. As shown in Figure 3D, the ratio of chlorophyll a/b in WT and lcd1 mutant leaves increased during storage, while LCD1 deletion caused the highest ratio compared with other groups. In contrast, the ratio of chlorophyll a/b in LCD1 overexpression almost remained unchanged. The above results indicate that the lcd1 mutation accelerated dark-induced leaf senescence and LCD1 overexpression delayed leaf yellowing and chlorophyll degradation.
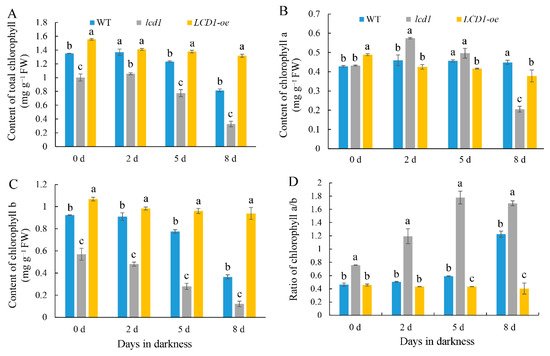

Figure 3. Changes in the contents of (A) total chlorophyll, (B) chlorophyll a, (C) chlorophyll b, and (D) the ratio of chlorophyll a/b in dark-stored detached leaves of 6-week-old wild-type (WT), lcd1, and LCD1-oe tomatoes for 0, 2, 5, and 8 days. Data are means of three biological replicates ± standard deviation (SD). Different letters above the columns stand for significant difference between two values (p < 0.05) at the same time point.
Leaf senescence is usually associated with the excessive accumulation of ROS; therefore, the levels of H2O2 and malondialdehyde (MDA) were monitored in wild-type, lcd1 mutant, and LCD1-oe leaves during dark-induced senescence. As shown in Figure 4A, there was no significant difference in H2O2 content between the different groups on day 0. During the dark-induced senescence, the H2O2 content in each group showed an increasing trend, of which the lcd1 group increased the fastest, followed by the wild-type and LCD1-oe group. However, H2O2 content in the LCD1-oe group increased slowly compared with other groups. As shown in Figure 4B, the change of MDA content among the groups also showed a similar trend to H2O2. The content of MDA in lcd1 leaves during storage was the highest compared with other groups, and the lowest MDA content was observed in LCD1-oe leaves. Therefore, it can be concluded that the overexpression of LCD1 could reduce the accumulation of ROS and MDA in leaves under dark-triggered senescence.
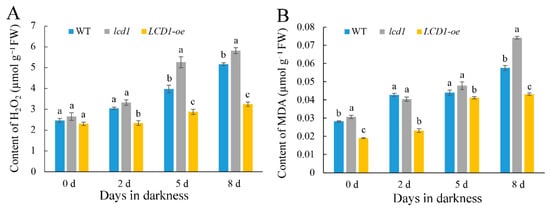

Figure 4. Changes in the contents of (A) H2O2 and (B) malondialdehyde (MDA) in dark-stored detached leaves of 6-week-old wild-type (WT), lcd1, and LCD1-oe tomatoes for 0, 2, 5, and 8 days. Data are means of three biological replicates ± standard deviation (SD). Different letters above the columns stand for significant difference between two values (p < 0.05) at the same time point.
5. Effect of LCD1 on the Expressions of Genes Related to Chlorophyll Degradation in Detached Tomato Leaves
Chlorophyll degradation marks the senescence stage of leaves. In order to explore the molecular mechanism of the differences in chlorophyll content of lcd1, LCD1-oe, and wild-type leaves during dark-induced senescence, the expression levels of key genes NYC1, PAO, PPH, and SGR1 in the chlorophyll degradation pathway were analyzed by RT-qPCR. The present data showed NYC1 was transcriptionally induced during dark-induced senescence in all groups (Figure 5A). In accordance with the early senescence phenotype of the lcd1 mutant and late senescence in LCD1-oe leaves, the expression of NYC1 was significantly higher in the lcd1 mutant and was less expressed in LCD1-oe leaves during dark storage. Three other genes—PAO (Figure 5B), PPH (Figure 5C), and SGR1 (Figure 5D)—were also analyzed at the transcriptional level in wild-type, lcd1 mutant, and LCD1-oe leaves during dark-induced senescence and similar changes to that of the NYC1 expression were observed. The higher expression of PAO, PPH, and SGR1 in lcd1 and lower expression in LCD1-oe again supported the role of LCD1 in delaying leaf senescence. The results suggest that LCD1 may delay the chlorophyll degradation by down-regulating the transcription of key genes in the chlorophyll degradation pathway.


Figure 5. Changes in the gene expressions of chlorophyll degradation related genes: (A) NYC1, (B) PAO, (C) PPH, and (D) SGR1 in detached leaves of 6-week-old wild-type (WT), lcd1, and LCD1-oe tomatoes stored in darkness for 0, 2, 5, and 8 days. Data are means of three biological replicates ± standard deviation (SD). Different letters above the columns stand for significant difference between two values (p < 0.05) at the same time point.
6. Effect of LCD1 on the Expressions of SAGs in Detached Tomato Leaves
To further analyze the senescence-alleviating role of LCD1, we conducted an RT-qPCR analysis to evaluate the expression patterns of senescence-associated genes (SAGs) in lcd1, LCD1-oe, and wild-type leaves during dark-induced senescence. As shown in Figure 6, SAG12, SAG15, and SAG113 were transcriptionally induced during dark-induced senescence. Compared with SAG15 and SAG113, SAG12 showed more fold changes during leaf senescence, which was 109.6 times in the wild type on day 8 compared with day 0 (Figure 6A). In accordance with the early senescence phenotype of the lcd1 mutant and late senescence in LCD1-oe leaves, the expression of the three SAGs was significantly higher in the lcd1 mutant and was less expressed in LCD1-oe leaves during dark storage, especially on day 8.
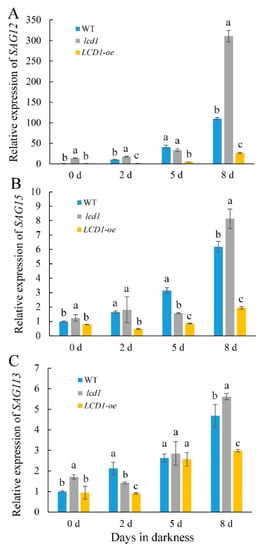

Figure 6. Changes in the gene expressions of senescence-related genes: (A) SAG12, (B) SAG15, and (C) SAG113 in detached leaves of 6-week-old wild-type (WT), lcd1, and LCD1-oe tomatoes stored in darkness for 0, 2, 5, and 8 days. Data are means of three biological replicates ± standard deviation (SD). Different letters above the columns stand for significant difference between two values (p < 0.05) at the same time point.
References
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Horti. 2021, 1, 5.
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136.
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NONYELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375.
- Chen, M. Chlorophyll modifications and their spectral extension in oxygenic photosynthesis. Annu. Rev. Biochem. 2014, 83, 317–340.
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131.
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160.
- Pruzinska, A.; Tanner, G.; Anders, I.; Roca, M.; Hortensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264.
- Schippers, J.H. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83.
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112.
- Gan, S.; Amasino, R.M. Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 1997, 113, 313–319.
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328.
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.J.; Gan, S.S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812.
- Wei, B.; Zhang, W.; Chao, J.; Zhang, T.; Zhao, T.; Noctor, G.; Liu, Y.; Han, Y. Functional analysis of the role of hydrogen sulfide in the regulation of dark-induced leaf senescence in Arabidopsis. Sci. Rep. 2017, 7, 2615.
- Kan, C.; Zhang, Y.; Wang, H.L.; Shen, Y.; Xia, X.; Guo, H.; Li, Z. Transcription factor NAC075 delays leaf senescence by deterring reactive oxygen species accumulation in Arabidopsis. Front. Plant Sci. 2021, 12, 634040.
- Wang, R. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol. Rev. 2012, 92, 791–896.
- Zhang, H.; Hu, L.Y.; Hu, K.D.; He, Y.D.; Wang, S.H.; Luo, J.P. Hydrogen sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. J. Integr. Plant Biol. 2008, 50, 1518–1529.
- Zhang, H.; Tang, J.; Liu, X.P.; Wang, Y.; Yu, W.; Peng, W.-Y.; Fang, F.; Ma, D.-F.; Wei, Z.-J.; Hu, L.Y. Hydrogen sulfide promotes root organogenesis in Ipomoea batatas, Salix matsudana and Glycine max. J. Integr. Plant Biol. 2009, 51, 1086–1094.
- Jin, Z.P.; Wang, Z.Q.; Ma, Q.X.; Sun, L.M.; Zhang, L.P.; Liu, Z.Q.; Liu, D.; Hao, X.; Pei, Y. Hydrogen sulfide mediates ion fluxes inducing stomatal closure in response to drought stress in Arabidopsis thaliana. Plant Soil 2017, 419, 141–152.
- Hu, K.D.; Zhang, X.Y.; Wang, S.S.; Tang, J.; Yang, F.; Huang, Z.Q.; Deng, J.Y.; Liu, S.Y.; Zhao, S.J.; Hu, L.Y.; et al. Hydrogen sulfide inhibits fruit softening by regulating ethylene synthesis and signaling pathway in tomato (Solanum lycopersicum). HortScience 2019, 54, 1824–1830.
- Yao, G.F.; Wei, Z.Z.; Li, T.T.; Tang, J.; Huang, Z.Q.; Yang, F.; Li, Y.H.; Han, Z.; Hu, F.; Hu, L.Y.; et al. Modulation of enhanced antioxidant activity by hydrogen sulfide antagonization of ethylene in tomato fruit ripening. J. Agric. Food Chem. 2018, 66, 10380–10387.
- Ni, Z.J.; Hu, K.D.; Song, C.B.; Ma, R.-H.; Li, Z.-R.; Zheng, J.-L.; Fu, L.-H.; Wei, Z.-J.; Zhang, H. Hydrogen sulfide alleviates postharvest senescence of grape by modulating the antioxidant defenses. Oxid. Med. Cell. Longev. 2016, 2016, 4715651.
- Ge, Y.; Hu, K.D.; Wang, S.S.; Hu, L.Y.; Chen, X.Y.; Li, Y.H.; Yang, Y.; Yang, F.; Zhang, H. Hydrogen sulfide alleviates postharvest ripening and senescence of banana by antagonizing the effect of ethylene. PLoS ONE 2017, 12, e0180113.
- Jin, Z.P.; Pei, Y.X. Physiological implications of hydrogen sulfide in plants: Pleasant exploration behind its unpleasant odour. Oxid. Med. Cell Longev. 2015, 2015, 397502.
- Álvarez, C.; Calo, L.; Romero, L.C.; Garcıá, I.; Gotor, C. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010, 152, 656–669.
- Jin, Z.; Sun, L.; Yang, G.; Pei, Y. Hydrogen sulfide regulates energy production to delay leaf senescence induced by drought stress in Arabidopsis. Front. Plant Sci. 2018, 9, 1722.
More
