Quantitative extraction of imaging features from medical scans (‘radiomics’) has become a major research topic in recent years. Numerous studies have emphasized the potential use of radiomics for computer-assisted diagnosis, as well as for predicting survival and response to treatment in patients with lung cancer. Furthermore, radiomics is appealing in that it enables full-field analysis of the lesion, provides nearly real-time results, and is non-invasive.
- PET/CT
- Texture
- Shape
- Radiomics
- Machine learning
- Lung cancer
1. Introduction
Lung cancer is the second most common type of cancer in men and women worldwide, with an estimated lifetime prevalence of about 1/15 and 1/17, respectively, for the two genders[1]. In Italy, there were ≈42,500 new cases in 2019, accounting for ≈11% of all the newly diagnosed cancers in the same year[2]. Five-year survival rates of patients with lung cancer vary considerably depending on the type and stage of the disease, and they range between a dismal 3% for distant small-cell lung cancer (SCLC) to 60% for localized non-small-cell lung cancer (NSCLC)[3]. Timely detection and correct management are therefore essential to improve the clinical outcome of patients with lung cancer.
In the last few years, computerized analysis of 3D scans from Computed Tomography (CT), Positron Emission Tomography (PET), and Magnetic Resonance Imaging (MRI) has received a great deal of attention as a means to improve the clinical management of a number of disorders. It is believed that radiomics has the potential to improve on traditional, manual interpretation by detecting features and patterns that otherwise would go unnoticed to the human eye[4][5]. By leveraging on large datasets (hence the suffix ‘-omics’) and artificial intelligence techniques, radiomics may help predict the type of disease, survival, and response to therapy[6][7]. There are also a number of logistic advantages in this approach, such as providing nearly real-time results and not requiring any invasive procedure for the patient. Furthermore, compared with standard biopsy, radiomics can offer not only a full-field analysis of one lesion but also of more lesions within the examined area, and, depending on the protocol used, of the whole body too[8].
Fluorine 18 (18F) fluorodeoxyglucose Positron Emission Tomography–Computed Tomography (PET/CT) is nowadays the mainstay in the management of lung cancer, having greatly improved patient diagnosis, staging, and follow-up [9]. The role of radiomics in this context has therefore attracted increasing interest in recent times[10][11][12][13][14]
2. Methodology
The overall workflow in radiomics involves six well-defined steps as described below.
2.1 Acquisition
This is the procedure whereby the scans are obtained, and includes both the examination itself and the patient preparation protocol. The output is a three-dimensional matrix of intensity values (voxel model), which in the remainder we refer to as the raw data. A wide range of parameters intervene in the acquisition process, among them tube current and voltage (for CT); spatial resolution (voxel size), reconstruction algorithm and related settings both for CT and PET. All these variables may have a significant impact on the radiomics features computed
, with certain features being affected more than others
[16]
.
2.2. Pre-Processing
Pre-processing may involve spatial filtering, windowing, and/or resampling. The objective of the first can be either to reduce noise or emphasize features at different scales. Common tools for this task are Butterworth smoothing[17], Gaussian filters [18], and Laplacian of Gaussian filters[19]. Windowing consists of applying a lower and upper threshold to the intensity values of the raw data, this way defining a range of acceptable values. Resampling amounts to changing the number of bits used for the encoding, which is commonly 12 or 16 for the raw data. This is typically reduced to eight, six, or four before feature extraction [17][20][21]. Pre-processing is a crucial step in the workflow and may significantly affect the overall outcome, as numerous experiments have demonstrated[16][17].
2.3. Segmentation
Segmentation (delineation) is the process whereby the part of the scan that is relevant for the analysis (Region of Interest—ROI) is separated from the background. The output of this step is a binary (boolean) matrix the same size of the raw data, where ‘true’ (1) indicates that the voxel belongs to the ROI, ‘false’ (0), otherwise. Segmentation is a time-consuming and rather complex step, for many lesions will show unclear and ill-defined borders. The process is also complicated by the presence of areas such as necrosis, atelectasis, and/or inflammation, whose role in the radiomics work-flow is not fully understood yet. Although a number of automated (e.g., adaptive thresholding[22], convolutional networks) and semi-automated (e.g., level-set[23], region growing[24]) methods have been proposed, manual delineation is still regarded by many as the gold standard[6].
2.4. Feature Extraction
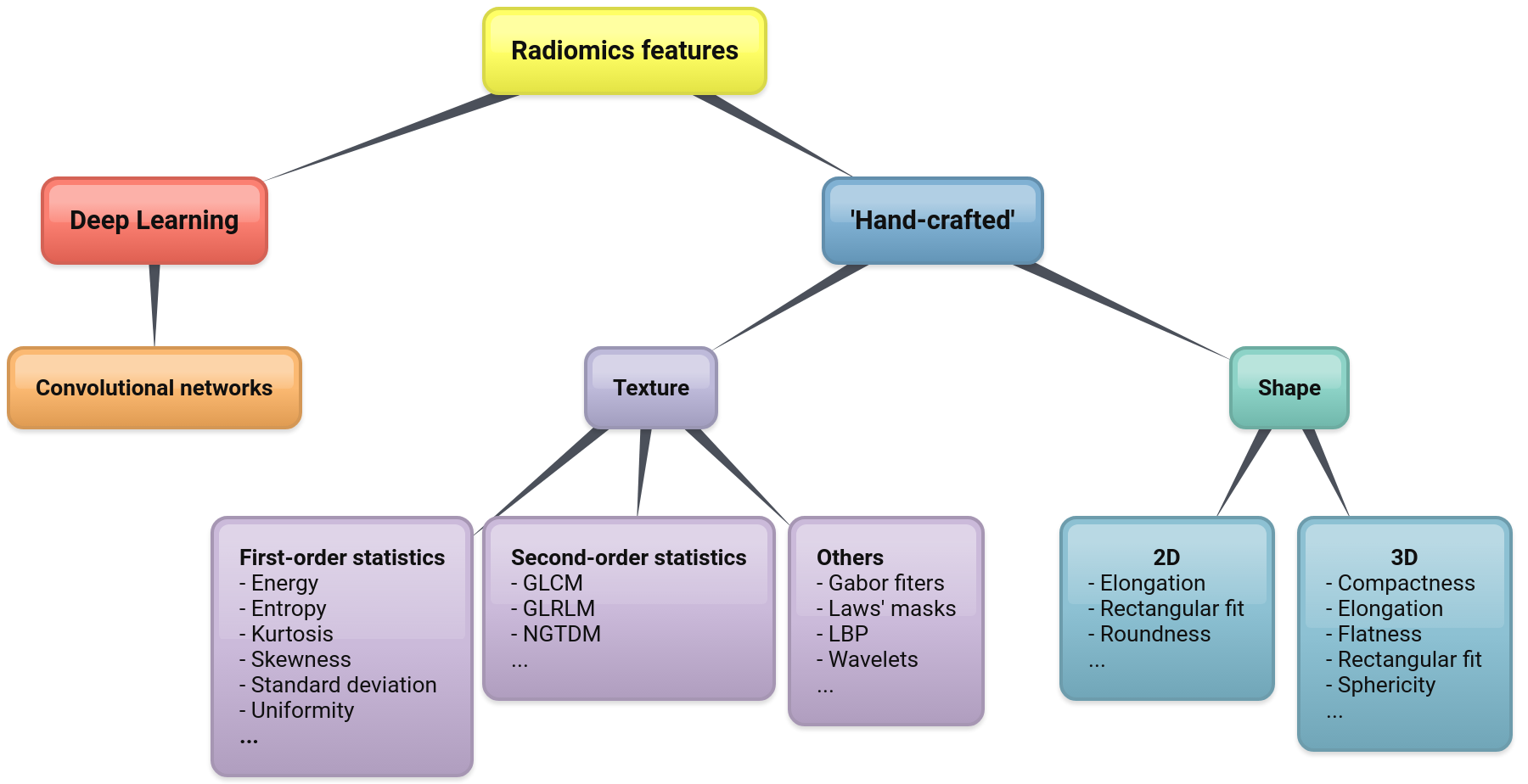
Feature extraction is a pivotal step in the whole procedure and involves computing a set of quantitative parameters (image features or, simply, features) from the region of interest. The features should obviously correlate with the endpoint of the clinical condition investigated. At present, there are two main classes of features: the ‘hand-designed’ (or ‘hand-crafted’) ones and those based on Deep Learning (see Figure 1 for a possible taxonomy). Hand-crafted features are obtained via some suitable mathematical functions that are essentially designed by hand (hence the name). Most common among them are shape and texture features[25]. By contrast, Deep Learning features are obtained implicitly by training on large datasets of images.
Figure 1: A taxonomy of radiomics features
2.5. Post Processing
Post-processing consists of transforming the features through some suitable procedures, the most common being feature selection and feature generation. Feature selection consists of retaining a subset of the original features by selecting the most discriminative ones. This is crucial in radiomics, for some image features tend to be strongly correlated with one another[26]. Approaches to feature selection come in different varieties, such as correlation-based selection, reduction based on mutual information gain, recursive elimination, and Lasso regularization (see[27] for a review on this). Feature generation involves obtaining new features by combining of the original ones through some suitable transformations, such as Linear Discriminant Analysis (LDA), Principal Component Analysis (PCA), and Multi-Dimensional Scaling (MDS)[28][29].
2.6. Data Analysis
Data analysis comprises two separate steps: the first (model building), in which a classification and/or regression model is generated; the second, where the model is used to make predictions about the case or cohort of patients under evaluation. Model building involves (a) establishing the type of classifier or regressor to be used, and (b) feeding the model with a set of pre-classified cases—i.e., arrays of features/label pairs where the label indicates the clinical condition of the corresponding subject. This process of presenting the model with pre-classified cases is usually referred to as training. Crucial to this step, of course, is the availability of large enough datasets of pre-classified cases (ground truth).
3. Applications
Here below we present four typical applications of PET/CT radiomics in lung cancer.
3.1. Discrimination between Benign and Malignant Pulmonary Nodules
Solitary pulmonary nodules (SPN) are relatively common findings in the clinical practice, although the available data about the estimated prevalence at CT examination vary significantly[30][31]. Clinical management of SPN poses significant challenges, for a non-negligible fraction of them (estimated between 3.7% and 5.5%[32]) may represent malignant lesions. Traditionally, the evaluation involved manual assessment of some key image characteristics at CT that are considered strong indicators of benignity or malignancy[33]. In this scenario, recent studies have shown that prediction models based on quantitative imaging features can help differentiate between benign, malignant, and inflammatory pulmonary nodules[34][35][36][37].
3.2. Classification between Primary and Metastatic Lesions; Histological Subtyping
Detailed lesion characterisation has important implications for the management of patients with lung cancer. Differential diagnosis between primary and metastatic lesions, for instance, is crucial for stratification as well as for establishing the optimal treatment strategy[38]. Likewise, the correct identification of histological subtype has a strong influence on the outcome and determination of the most appropriate therapy[39][40].
3.3. Prediction of Survival
Prediction of survival plays an important role for triaging and, consequently, for determining the suitability of subjects for different treatment options. Survival studies, however, are notoriously difficult due to the long follow-up required and the presence of many confounding factors. It is not surprising then that results in this topic are less clear-cut than in the other potential applications[21].
3.4. Prediction of Response to Treatment
Predicting response to treatment—and in particular to chemotherapy, radiotherapy, and immunotherapy—is crucial to maximize the outcome, and, at the same time, minimize the side effects by avoiding the administration of inefficient treatments[19]. In previous studies textural features from PET/CT scans at the baseline were found to correlate with local recurrence and disease-specific survival in patients treated with radiotherapy[9][20]. Radiomic signatures from baseline PET/CT also proved useful to predict disease-free survival in NSCLC patients undergoing surgery[41].
4. Conclusions
Quantitative image analysis of PET/CT scans has attracted increasing research interest for the management of patients with lung cancer. There is growing evidence that radiomics can provide an added value to standard imaging analysis tools such as volume, SUV and density. However, the field is still at an early stage and further work is required to confirm the benefits and potential advantages in the clinical practice.
References
- Key Statistics for Lung Cancer. 2019 . American Cancer Society. Retrieved 2020-8-27
- I Numeri del Cancro in Italia. 2019 . Associazione Italiana di Oncologia Medica; Associazione Italiana dei Registri Tumori. Retrieved 2020-8-27
- Lung Cancer Survival Rates. 2019. . American Cancer Society. Retrieved 2020-8-27
- Robert J. Gillies; P E Kinahan; Hedvig Hricak; Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563-577, 10.1148/radiol.2015151169.
- Simon A Keek; Ralph Th Leijenaar; Arthur Jochems; Henry C Woodruff; A review on radiomics and the future of theranostics for patient selection in precision medicine.. The British Journal of Radiology 2018, 91, 20170926, 10.1259/bjr.20170926.
- Stefania Rizzo; Francesca Botta; Sara Raimondi; Daniela Origgi; Cristiana Fanciullo; Alessio Giuseppe Morganti; Massimo Bellomi; Radiomics: the facts and the challenges of image analysis. European Radiology Experimental 2018, 2, 36, 10.1186/s41747-018-0068-z.
- Marco Aiello; Carlo Cavaliere; Antonio D’Albore; Marco Salvatore; The Challenges of Diagnostic Imaging in the Era of Big Data. Journal of Clinical Medicine 2019, 8, 316, 10.3390/jcm8030316.
- Isabella Castiglioni; Maria Carla Gilardi; Radiomics: is it time to compose the puzzle?. Clinical and Translational Imaging 2018, 6, 411-413, 10.1007/s40336-018-0302-y.
- Anastasia Oikonomou; Farzad Khalvati; Pascal N Tyrrell; Masoom A. Haider; Usman Tarique; Laura Jimenez-Juan; Michael C. Tjong; Ian Poon; Armin Eilaghi; Lisa Ehrlich; et al.Patrick Cheung Radiomics analysis at PET/CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy. Scientific Reports 2018, 8, 1-11, 10.1038/s41598-018-22357-y.
- Geewon Lee; Ho Yun Lee; Hyunjin Park; Mark L. Schiebler; Edwin J.R. Van Beek; Yoshiharu Ohno; Joon Beom Seo; Ann Leung; Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. European Journal of Radiology 2017, 86, 297-307, 10.1016/j.ejrad.2016.09.005.
- Bojiang Chen; Rui Zhang; Yuncui Gan; Lan Yang; Weimin Li; Development and clinical application of radiomics in lung cancer. Radiation Oncology 2017, 12, 154, 10.1186/s13014-017-0885-x.
- Rajat Thawani; Michael McLane; Niha G. Beig; Soumya Ghose; Prateek Prasanna; Vamsidhar Velcheti; Anant Madabhushi; Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018, 115, 34-41, 10.1016/j.lungcan.2017.10.015.
- Cameron Hassani; Bino A. Varghese; Jorge Nieva; Vinay Duddalwar; Radiomics in Pulmonary Lesion Imaging. American Journal of Roentgenology 2019, 212, 497-504, 10.2214/ajr.18.20623.
- Clément Bailly; Caroline Bodet-Milin; Mickaël Bourgeois; Sébastien Gouard; Catherine Ansquer; Matthieu Barbaud; Jean-Charles Sébille; Michel Chérel; Françoise Kraeber-Bodere; Thomas Carlier; et al. Exploring Tumor Heterogeneity Using PET Imaging: The Big Picture. Cancers 2019, 11, 1282, 10.3390/cancers11091282.
- Floris H. P. Van Velden; G. M. Kramer; Virginie Frings; Ida A. Nissen; Emma R. Mulder; Adrianus J. De Langen; Otto S. Hoekstra; Egbert F. Smit; Ronald Boellaard; Repeatability of Radiomic Features in Non-Small-Cell Lung Cancer [(18)F]FDG-PET/CT Studies: Impact of Reconstruction and Delineation.. Molecular Imaging and Biology 2016, 18, 788-95, 10.1007/s11307-016-0940-2.
- Alberto Traverso; Leonard Wee; Aldo Dekker; Robert Gillies; Repeatability and Reproducibility of Radiomic Features: A Systematic Review.. International Journal of Radiation Oncology*Biology*Physics 2018, 102, 1143-1158, 10.1016/j.ijrobp.2018.05.053.
- Xenia Fave; Lifei Zhang; Jinzhong Yang; Dennis Mackin; Peter Balter; Daniel Gomez; David Followill; A. Kyle Jones; Francesco C. Stingo; Laurence E. Court; et al. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Translational Cancer Research 2016, 5, 349-363, 10.21037/tcr.2016.07.11.
- Luca Brunese; Barbara Greco; Francesca Rosa Setola; Francesco Lassandro; Mario Rosario Guarracino; Marialuisa De Rimini; Sergio Piccolo; Nicolina De Rosa; Roberto Muto; Andrea Bianco; et al.Pietro MutoRoberto GrassiAntonio Rotondo Studio comparativo con esame TC contrastografico e con PET-TC del tumore polmonare non a piccole cellule. Medical Science Monitor 2013, 19, 95-101, 10.12659/MSM.883759.
- M. Ravanelli; D. Farina; Mauro Morassi; Elisa Roca; Giuseppe Cavalleri; Gianfranco Tassi; Roberto Maroldi; Texture analysis of advanced non-small cell lung cancer (NSCLC) on contrast-enhanced computed tomography: prediction of the response to the first-line chemotherapy. European Radiology 2013, 23, 3450-3455, 10.1007/s00330-013-2965-0.
- Thomas Pyka; Ralph Bundschuh; Nicolaus Andratschke; Benedikt Mayer; Hanno M Specht; Laszlo Papp; Norbert Zsótér; Markus Essler; Textural features in pre-treatment [F18]-FDG-PET/CT are correlated with risk of local recurrence and disease-specific survival in early stage NSCLC patients receiving primary stereotactic radiation therapy. Radiation Oncology 2015, 10, 1-9, 10.1186/s13014-015-0407-7.
- Francesco Bianconi; Mario Luca Fravolini; Raquel Bello-Cerezo; Matteo Minestrini; Michele Scialpi; Barbara Palumbo; Evaluation of Shape and Textural Features from CT as Prognostic Biomarkers in Non-small Cell Lung Cancer. Anticancer Research 2018, 38, 2155-2160, 10.21873/anticanres.12456.
- Chithra, A. and Renjen Roy, R. Otsu’s Adaptive Thresholding Based Segmentation for Detection of LungNodules in CT Image. In Proceedings of the 2nd International Conference on Trends in Electronics andInformatics (ICOEI), Tirunelveli, India, 11–12 May 2018; pp. 1303–1307
- Karthik Krishnan; Luis Ibanez; Wesley Turner; Julien Jomier; Ricardo S. Avila; An open-source toolkit for the volumetric measurement of CT lung lesions.. Optics Express 2010, 18, 15256-15266, 10.1364/oe.18.015256.
- Stephen Sf Yip; Chintan Parmar; Daniel Blezek; Raúl San José Estépar; Steve Pieper; John Kim; Hugo Aerts; Application of the 3D slicer chest imaging platform segmentation algorithm for large lung nodule delineation. PLoS ONE 2017, 12, e0178944, 10.1371/journal.pone.0178944.
- Francesco Bianconi; Mario Luca Fravolini; Isabella Palumbo; Barbara Palumbo; Shape and Texture Analysis of Radiomic Data for Computer-Assisted Diagnosis and Prognostication: An Overview. Lecture Notes in Mechanical Engineering 2019, 1, 3-14, 10.1007/978-3-030-31154-4_1.
- Francesco Bianconi; Isabella Palumbo; Mario Luca Fravolini; Rita Chiari; Matteo Minestrini; Luca Brunese; Barbara Palumbo; Texture Analysis on [18F]FDG PET/CT in Non-Small-Cell Lung Cancer: Correlations Between PET Features, CT Features, and Histological Types. Molecular Imaging and Biology 2019, 21, 1200-1209, 10.1007/s11307-019-01336-3.
- Beatriz Remeseiro; Verónica Bolón-Canedo; A review of feature selection methods in medical applications.. Computers in Biology and Medicine 2019, 112, 103375, 10.1016/j.compbiomed.2019.103375.
- Van der Heijden, F.; Duin, R.P.W.; de Ridder, D.; Tax, D.M.J.. Classification, Parameter Estimation and State Estimation. An Engineering Approach Using Matlab; John Wiley & Sons: Chichester, UK, 2004; pp. 440.
- Theodoridis, S.; Koutroumbas, K.. Pattern Recognition, 3rd ed.; Academic Press: Cambridge, MA, USA, 2006; pp. 856.
- Gary Hammerschlag; Jingyi Cao; Kellie Gumm; Louis Irving; Daniel Steinfort; The Prevalence of Incidental Pulmonary Nodules on Computed Tomography of the Thorax in Trauma Patients. Internal Medicine Journal 2015, 45, 630-633, 10.1111/imj.12755.
- Émilie Marrer; Damien Jolly; Patrick Arveux; C. Lejeune; Marie-Christine Woronoff-Lemsi; Jérémie Jegu; Francis Guillemin; Michel Velten; Incidence of solitary pulmonary nodules in Northeastern France: a population-based study in five regions.. BMC Cancer 2017, 17, 47, 10.1186/s12885-016-3029-z.
- Annette McWilliams; C. Martin Tammemagi; John R. Mayo; Heidi Roberts; Geoffrey Liu; Kam Soghrati; Kazuhiro Yasufuku; Simon Martel; Francis Laberge; Michel Gingras; et al.Sukhinder Atkar-KhattraChristine D. BergKen EvansRichard FinleyJohn YeeJohn EnglishPaola NasuteJohn GoffinSerge PuksaLori StewartScott TsaiMichael R. JohnstonDaria ManosGarth NicholasGlenwood D. GossJean M. SeelyKayvan AmjadiAlain TremblayPaul BurrowesPaul MacEachernRick BhatiaMing TsaoStephen Lam Probability of cancer in pulmonary nodules detected on first screening CT.. New England Journal of Medicine 2013, 369, 910-9, 10.1056/NEJMoa1214726.
- A.J. Edey; D.M. Hansell; Incidentally detected small pulmonary nodules on CT. Clinical Radiology 2009, 64, 872-884, 10.1016/j.crad.2009.03.006.
- Shiteng Suo; Jiejun Cheng; Mengqiu Cao; Qing Lu; Yan Yin; Jianrong Xu; Hua-Wei Wu; Assessment of Heterogeneity Difference Between Edge and Core by Using Texture Analysis. Academic Radiology 2016, 23, 1115-1122, 10.1016/j.acra.2016.04.009.
- José Raniery Ferreira Júnior; Marcelo Costa Oliveira; Paulo Mazzoncini De Azevedo Marques; Characterization of Pulmonary Nodules Based on Features of Margin Sharpness and Texture. Journal of Digital Imaging 2017, 31, 451-463, 10.1007/s10278-017-0029-8.
- Wei Wu; Larry A. Pierce; Yuzheng Zhang; Sudhakar N. J. Pipavath; Timothy W. Randolph; Kristin J. Lastwika; Paul D. Lampe; A. McGarry Houghton; Haining Liu; Liming Xia; et al.P E Kinahan Comparison of prediction models with radiological semantic features and radiomics in lung cancer diagnosis of the pulmonary nodules: a case-control study. European Radiology 2019, 29, 6100-6108, 10.1007/s00330-019-06213-9.
- Yoganand Balagurunathan; Matthew B. Schabath; Hua Wang; Ying Liu; Robert J. Gillies; Quantitative Imaging features Improve Discrimination of Malignancy in Pulmonary nodules. Scientific Reports 2019, 9, 8528, 10.1038/s41598-019-44562-z.
- Dani S. Zander; Primary vs Metastatic Pulmonary Adenocarcinoma. Chest 2010, 137, 3-4, 10.1378/chest.09-1514.
- Takayuki Fukui; Tetsuo Taniguchi; Koji Kawaguchi; Koichi Fukumoto; Shota Nakamura; Yukinori Sakao; Kohei Yokoi; Comparisons of the clinicopathological features and survival outcomes between lung cancer patients with adenocarcinoma and squamous cell carcinoma. General Thoracic and Cardiovascular Surgery 2015, 63, 507-513, 10.1007/s11748-015-0564-5.
- Silvia Rinaldi; Rossana Berardi; Lung cancer prognosis: can histological patterns and morphological features have a role in the management of lung cancer patients?. Annals of Translational Medicine 2017, 5, 353-353, 10.21037/atm.2017.05.18.
- Margarita Kirienko; Luca Cozzi; Lidija Antunovic; Lisa Lozza; A. Fogliata; Emanuele Voulaz; Alexia Rossi; Arturo Chiti; Martina Sollini; Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. European Journal of Pediatrics 2017, 45, 207-217, 10.1007/s00259-017-3837-7.