Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Beatrix Zheng and Version 1 by Eric Kalkhoven.
Tribbles proteins play various roles in cancer initiation and progression. However, still little is known about their molecular actions. Here we developed a mass spectrometry-based approach to study the Tribbles interactomes, allowing us to discover new interactors and functions that might help to understand their behavior better. Our proteomics data highlight the ability of TRIB3 to interact with transcription regulatory proteins and point to a new role in gene repression. Systematic analyses like these will help to evaluate the potential of Tribbles proteins as biomarkers for disease diagnosis and prognosis.
- tribbles
- proteomics
- interactome
- breast cancer
1. Introduction
Kinases regulate a plethora of cellular processes and changes in their enzymatic activity are intimately linked to human diseases, hence the large research field studying the basic biology of kinases and their potential as therapeutic targets [1,2][1][2]. In addition to 518 kinases, the human genome also encodes ~60 pseudokinases, proteins that resemble serine/threonine and tyrosine protein kinases but lack several amino acids critical for enzymatic activity [3,4][3][4]. The human pseudokinase family includes the three members of the Tribbles (TRIB) family—TRIB1, TRIB2 and TRIB3—that share a high degree of homology as well as similar domain compositions [5,6][5][6]. They can be divided into three major domains: an N-terminal domain, associated with protein stability and subcellular localization [7,8][7][8]; a well-conserved, centrally located pseudokinase domain; and a C-terminal domain, wherein the binding motifs of MAPK and COP1 are found [9,10][9][10]. A fourth, more distally related protein, called STK40, shares important similarities in terms of function and structure [11,12][11][12].
Tribbles proteins have been implicated in multiple critical signaling and metabolic processes and alterations in their expression and/or activity is linked to various human diseases [13]. While lacking intrinsic enzymatic activity, Tribbles proteins exert their biological roles predominantly via binding to other proteins, including kinases, phosphatases, transcription factors and components of the ubiquitin-proteosome system [14,15,16][14][15][16]. This diverse range of interactors explains, at least in part, the difficulties to associate a TRIB family member with a single specific cellular pathway or role. In addition, it should be noted that different and even contradictory observations have been made regarding the subcellular localization of Tribbles proteins, suggesting their localization and thereby function depends on cellular context and conditions [17,18][17][18].
In recent years many studies have pointed to Tribbles proteins as important modulators of cancer initiation and progression [19,20,21,22][19][20][21][22]. Therefore, Tribbles proteins hold potential as biomarkers of disease diagnosis and prognosis as well as pharmaceutical targets for a number of cancers [23]. For example, TRIB1 upregulation is significantly associated with metastasis and poor prognosis in prostate cancer [24], it has been shown that TRIB1 mediates radioresistance in glioma cells by an HDAC1-dependent pathway [25] and high levels of TRIB1 are associated with poor breast cancer survival through the regulation of PI3K-NFκB pathway [26]. TRIB2 has been shown to contribute to tumorigenesis in lung cancer through the downregulation of C/EBPα [27] and TRIB2 direct interaction with AKT has been shown to be an important mechanism that contributes to resistance to anti-cancer drug therapy [28]. Finally, TRIB3 has been shown to support breast and colorectal cancer stemness through the interaction with AKT and beta-catenin respectively [29,30][29][30]. These examples illustrate that the different Tribbles family members can all play a regulatory role in cancer initiation and progression, but their contribution may be tumor type specific. Furthermore, these examples also add to anecdotal evidence that critical interacting proteins may differ between Tribbles family members and to previous reports that the affinities of distinct TRIB proteins to the same binding partner may differ [31]. We hypothesize, therefore, that particular Tribbles functions are dictated by its interactome—the specific set of proteins with which a tribble family member is interacting within a given biological setting—and that improving our understanding of how these interactions take place will help to define the roles of Tribbles proteins in each context. To date, comprehensive Tribbles interactomes have not been reported.
Mass spectrometry (MS)-based proteomics approaches have been widely used in recent decades to study and identify protein–protein interactions (PPIs) [32]. Affinity-purification mass spectrometry (AP–MS) is used for the purification of a protein (endogenous or tagged) and its interacting partners from a cell lysate [32]. This technique relies on the affinity of an antibody (or nanobody) for a protein and is followed by MS analysis [32]. We have previously used this approach successfully to identify the interactomes of various intracellular proteins [33,34,35][33][34][35].
2. Current Insights
Pseudokinases, such as the three human TRIB proteins, hold promise as biomarkers in cancer, but their molecular functions are still incompletely understood. Here we reported a systematic characterization of TRIB1, -2 and -3 interactomes in HEK 293T cells to provide a better understanding of the differences and redundancies in Tribbles’ functions. In addition, our mass spectrometry-based approach revealed the importance of the intrinsically disordered N-Terminal domain of TRIB3 in the interaction with transcriptional regulatory proteins. We showed that TRIB3 is associated with transcriptional repression and that this role is mostly carried by the N-Terminal of TRIB3. Moreover, we discover new interactors of TRIB1 and -3 in breast cancer cells that might help to understand the role of these proteins in cancer pathophysiology.
The study of the function of pseudoenzymes presents obvious difficulties in comparison with their enzymatically active counter partners, as no catalytic product can be measured as a read-out of their activity. Most of these pseudoenzymes rely on protein–protein interactions (PPI) to exert their function and several mass spectrometry-based techniques can be used for the identification of interactors, such as proximity ligation or crosslinking mass spectrometry, all with particular advantages and disadvantages [32]. Our data shows how powerful is the use of AP–MS approaches for the discovery of new interactors and the study of pseudoenzyme function. Modern mass spectrometers have a tremendous sensitivity that allows them to detect the smallest contaminant and, therefore, a quantitative filter must be introduced to differentiate between genuine interactors and background noise. These quantitative filters can be introduced in the form of isotopes or in the form of algorithms for label-free quantification; an example of the last is the intensity-based absolute quantification (iBAQ) used in this study, which allowed us to determine protein abundance. Taken together, this shows that quantitative MS-based proteomics is the most powerful method for identifying PPI and studying pseudoenzyme function to date.
Through AP–MS we confirmed previously reported Tribbles-interacting proteins and identified novel partners. As demonstrated before, we show that all three human tribbles family members can interact with the E3 ubiquitin ligase COP1. However, it seems that TRIB1 function is more dominated by the interaction with COP1, and that explains the high amount of proteasomal regulatory proteins as well as the low abundance of other interactors. TRIB2 and -3 also interact with COP1 but, next to these, many other interactors, not related to proteasomal degradation, were detected. This also seems the case when we compared the interactomes of TRIB1 and -3 in breast cancer cells. Moreover, TRIB1 protein expression was lower when compared to TRIB3 upon induction with doxycycline (Figure 31B), and, thus, could be reverted when proteasomal degradation was inhibited (data not shown), indicating that the lower amount of TRIB1 protein was the result of proteasomal degradation and was not due to different responses to doxycycline induction. TRIB1 subcellular localization appeared to be mostly cytoplasmatic in comparison with TRIB3, which showed predominant nuclear localization; this can also explain the difference in protein stability and interactomes. In addition, our data also demonstrates some interactions that had been suggested in literature before but not experimentally demonstrated, such as the interaction with p53 or the interaction with CDKN1 [72,73][36][37]. These interactions might be related to the ability of Tribbles to regulate the cell cycle and therefore the implications for cancer research are potentially very important. Both of these proteins are among the most commonly found mutated across all cancer types [74,75][38][39]. These interactions, together with others described above, might suggest a role of Tribbles in DNA damage. Furthermore, whether for example the TRIB3–CDKN1 interaction contributes to the increased proliferation observed upon long-term overexpression of TRIB3 in MCF7 cells [62][40] remains to be established.
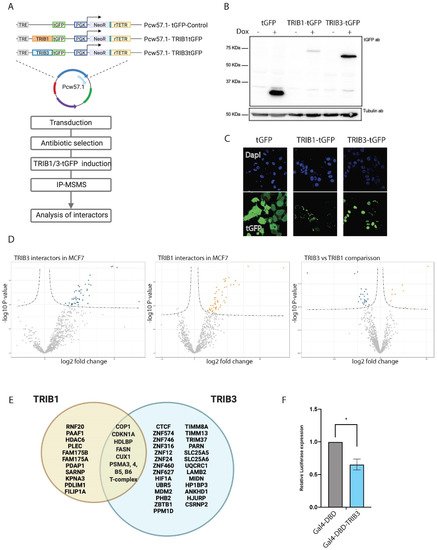

Figure 1. TRIB1 and TRIB3 interactors in MCF7 cells. (A) Schematic representation of inducible constructs and workflow of the AP–MS experiments followed in MCF7 cells. (B) Western blot using t-GFP antibody sowing inducible expression of TRIB1-tGFP and TRIB3-tGFP upon doxycycline treatment and Tubulin expression as loading control. (C) Confocal images taken at 40× magnification showing tGFP, TRIB1-tGFP and TRIB3-tGFP localization upon doxycycline induction. (D) Volcano plots of TRIB1 and TRIB3 interactors compared with tGFP control in MCF7 cells and a volcano plot showing the comparison between TRIB1 and TRIB3 interactome in these cells. (E) Venn diagram of similar and different interactors between TRIB1 and TRIB3 in MCF7 cells detected in the AP–MSMS experiments. (F) Gal4 reporter assay of Gal4-DBD and Gal4-TRIB3 in MCF7 cells. Data is normalized using Renilla luciferase. Data is indicated as mean ± SEM. p-values were calculated using two-tailed Student’s t-test (* p < 0.05).
Finally, we report many novel interacting proteins that interact with one or more Tribbles family members. Amongst the cellular proteins with well-established functions is the metabolic enzyme FASN. Interestingly, FASN is a major regulator of neoplastic lipogenesis and is commonly found overexpressed in many cancers [65][41], is a metabolic oncogene that has been suggested as an attractive target for cancer therapy [76][42] and, given the ability of TRIB1 and -3 to mark proteins for proteasomal degradation, this interaction represents a promising therapeutic approach for breast cancer. The class of well-characterized TRIB3 interacting proteins also includes the transcriptional repressors ZBTB1 and SPEN, which may be responsible for the transcriptional repression observed when TRIB3 is tethered to DNA. The ability of tribbles to regulate the functions of transcription factors has been reported before [29,30,31][29][30][31]; however, their role as a transcriptional repressor has not been shown before. While novel Tribbles interacting proteins with well-characterized functions may present immediate new entries for future research, interacting proteins with poorly understood functions such as the zinc finger proteins found interacting with TRIB3, obviously will need more characterization before their value—be it therapeutic or more fundamental—can be assessed. It should be noted that the protein kinase AKT/PKB, a previously described interaction partner of TRIB3 [14], was detected in the TRIB1 interactome but was not a dominant hit in the TRIB3 interactomes in either HEK293T or MCF7 cells. Furthermore, when we compared the TRIB3 interactomes between the genetic variants R84 and Q84—harboring an arginine and glutamine residue at position 84, respectively—no significant differences in interactomes were found (data not shown). The R variant was reported to be a more potent inhibitor of insulin signaling through stronger AKT binding when tested in hepatocytes [77][43]. Together, these findings support the view that TRIB interactomes may harbor a uniform component (overlap between for example HEK293T cells and MCF7 cells) as well as a flexible component that depends on cell type (e.g., hepatocyte vs. HEK293T cells vs. MCF7 breast cancer cells) or cellular status (e.g., proliferative status, metabolic status). Analyzing and comparing TRIB interactomes in more cell types and under different conditions is, therefore, an important future direction.
References
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015, 172, 2675–2700.
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569.
- Byrne, D.P.; Foulkes, D.M.; Eyers, P.A. Pseudokinases: Update on their functions and evaluation as new drug targets. Future Med. Chem. 2017, 9, 245–265.
- Richmond, L.; Keeshan, K. Pseudokinases: A tribble-edged sword. FEBS J. 2020, 287, 4170–4182.
- Eyers, P.A.; Keeshan, K.; Kannan, N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol. 2017, 27, 284–298.
- Kiss-Toth, E.; Velasco, G.; Pear, W.S. Tribbles at the cross-roads. Biochem. Soc. Trans. 2015, 43, 1049–1050.
- Soubeyrand, S.; Martinuk, A.; Lau, P.; McPherson, R. TRIB1 Is Regulated Post-Transcriptionally by Proteasomal and Non-Proteasomal Pathways. PLoS ONE 2016, 11, e0152346.
- Wang, J.; Zhang, Y.; Weng, W.; Qiao, Y.; Ma, L.; Xiao, W.; Yu, Y.; Pan, Q.; Sun, F. Impaired phosphorylation and ubiquitination by p70 S6 kinase (p70S6K) and Smad ubiquitination regulatory factor 1 (Smurf1) promote tribbles homolog 2 (TRIB2) stability and carcinogenic property in liver cancer. J. Biol. Chem. 2013, 288, 33667–33681.
- Murphy, J.; Nakatani, Y.; Jamieson, S.A.; Dai, W.; Lucet, I.S.; Mace, P.D. Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase. Structure 2015, 23, 2111–2121.
- Yokoyama, T.; Kanno, Y.; Yamazaki, Y.; Takahara, T.; Miyata, S.; Nakamura, T. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood 2010, 116, 2768–2775.
- Durzynska, I.; Xu, X.; Adelmant, G.; Ficarro, S.B.; Marto, J.A.; Sliz, P.; Uljon, S.; Blacklow, S.C. STK40 Is a Pseudokinase that Binds the E3 Ubiquitin Ligase COP1. Structure 2017, 25, 287–294.
- Yu, H.; He, K.; Wang, L.; Hu, J.; Gu, J.; Zhou, C.; Lu, R.; Jin, Y. Stk40 represses adipogenesis through translational control of CCAAT/enhancer-binding proteins. J. Cell Sci. 2015, 128, 2881–2890.
- Yokoyama, T.; Nakamura, T. Tribbles in disease: Signaling pathways important for cellular function and neoplastic transformation. Cancer Sci. 2011, 102, 1115–1122.
- Das, R.; Sebo, Z.; Pence, L.; Dobens, L.L. Drosophila tribbles antagonizes insulin signaling-mediated growth and metabolism via interactions with Akt kinase. PLoS ONE 2014, 9, e109530.
- Xiang, D.; Zhu, X.; Zhang, Y.; Zou, J.; Li, J.; Kong, L.; Zhang, H. Tribbles homolog 2 promotes hepatic fibrosis and hepatocarcinogenesis through phosphatase 1A-Mediated stabilization of yes-associated protein. Liver Int. 2021, 41, 1131–1147.
- Ferreira, B.I.; Santos, B.; Link, W.; De Sousa-Coelho, A.L. Tribbles pseudokinases in colorectal cancer. Cancers 2021, 13, 2825.
- Dobens, L.L.; Bouyain, S. Developmental roles of tribbles protein family members. Dev. Dyn. 2012, 241, 1239–1248.
- Kung, J.E.; Jura, N. The pseudokinase TRIB 1 toggles an intramolecular switch to regulate COP 1 nuclear export. EMBO J. 2019, 38, e99708.
- Yu, J.-J.; Zhou, D.-D.; Yang, X.-X.; Cui, B.; Tan, F.-W.; Wang, J.; Li, K.; Shang, S.; Zhang, C.; Lv, X.-X.; et al. TRIB3-EGFR interaction promotes lung cancer progression and defines a therapeutic target. Nat. Commun. 2020, 11, 3660.
- Shahrouzi, P.; Astobiza, I.; Cortazar, A.R.; Torrano, V.; Macchia, A.; Flores, J.M.; Niespolo, C.; Mendizabal, I.; Caloto, R.; Ercilla, A.; et al. Genomic and Functional Regulation of TRIB1 Contributes to Prostate Cancer Pathogenesis. Cancers 2020, 12, 2593.
- Kim, T.; Johnston, J.; Felipe, F.J.C.; Hamby, S.; Castillo-Lluva, S.; Consortium, T.C.; Goodall, A.H.; Velasco, G.; Ocana, A.; Muthana, M.; et al. TRIB1 regulates tumour growth via controlling tumour-associated macrophage phenotypes and is associated with breast cancer survival and treatment response. bioRxiv 2021.
- Stefanovska, B.; André, F.; Fromigué, O. Tribbles Pseudokinase 3 Regulation and Contribution to Cancer. Cancers 2021, 13, 1822.
- Ashton-Chess, J.; Giral, M.; Mengel, M.; Renaudin, K.; Foucher, Y.; Gwinner, W.; Braud, C.; Dugast, E.; Quillard, T.; Thebault, P.; et al. Tribbles-1 as a novel biomarker of chronic antibody-mediated rejection. J. Am. Soc. Nephrol. 2008, 19, 1116–1127.
- Lin, Z.-Y.; Huang, Y.-Q.; Zhang, Y.-Q.; Han, Z.-D.; He, H.-C.; Ling, X.-H.; Fu, X.; Dai, Q.-S.; Cai, C.; Chen, J.-H.; et al. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int. J. Cancer 2014, 135, 541–550.
- Tang, B.; Wu, W.; Zhang, Q.; Sun, Y.; Cui, Y.; Wu, F.; Wei, X.; Qi, G.; Liang, X.; Tang, F.; et al. Inhibition of tribbles protein-1 attenuates radioresistance in human glioma cells. Sci. Rep. 2015, 5, 15961.
- Gendelman, R.; Xing, H.; Mirzoeva, O.K.; Sarde, P.; Curtis, C.; Feiler, H.S.; McDonagh, P.; Gray, J.W.; Khalil, I.; Korn, W.M. Bayesian network inference modeling identifies TRIB1 as a novel regulator of cell-cycle progression and survival in cancer cells. Cancer Res. 2017, 77, 1575–1585.
- Grandinetti, K.B.; A Stevens, T.; Ha, S.; Salamone, R.J.; Walker, J.R.; Zhang, J.; Agarwalla, S.; Tenen, D.; Peters, E.C.; Reddy, A.V. Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPα. Oncogene 2011, 30, 3328–3335.
- Hill, R.; Kalathur, R.K.R.; Colaço, L.; Brandão, R.; Ugurel, S.; Futschik, M.; Link, W. TRIB2 as a biomarker for diagnosis and progression of melanoma. Carcinogenesis 2015, 36, 469–477.
- Yu, J.-M.; Sun, W.; Wang, Z.-H.; Liang, X.; Hua, F.; Li, K.; Lv, X.-X.; Zhang, X.-W.; Liu, Y.-Y.; Yu, J.-J.; et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 2019, 10, 5720.
- Hua, F.; Shang, S.; Yang, Y.W.; Zhang, H.Z.; Xu, T.L.; Yu, J.J.; Zhou, D.D.; Cui, B.; Li, K.; Lv, X.X.; et al. TRIB3 Interacts with β-Catenin and TCF4 to Increase Stem Cell Features of Colorectal Cancer Stem Cells and Tumorigenesis. Gastroenterology 2019, 156, 708.e15–721.e15.
- Guan, H.; Shuaib, A.; De Leon, D.D.; Angyal, A.; Salazar, M.; Velasco, G.; Holcombe, M.; Dower, S.K.; Kiss-Toth, E. Competition between members of the tribbles pseudokinase protein family shapes their interactions with mitogen activated protein kinase pathways. Sci. Rep. 2016, 6, 32667.
- Smits, A.H.; Vermeulen, M. Characterizing Protein-Protein Interactions Using Mass Spectrometry: Challenges and Opportunities. Trends Biotechnol. 2016, 34, 825–834.
- Smits, A.H.; Vermeulen, M. Quantitative liver proteomics identifies FGF19 targets that couple metabolism and proliferation. PLoS ONE 2017, 12, e0171185.
- Yuan, R.; Vos, H.; Van Es, R.M.; Chen, J.; Burgering, B.M.; Westendorp, B.; De Bruin, A. Chk1 and 14-3-3 proteins inhibit atypical E2Fs to prevent a permanent cell cycle arrest. EMBO J. 2018, 37, e97877.
- Van Nuland, R.; Smits, A.H.; Pallaki, P.; Jansen, P.W.T.C.; Vermeulen, M.; Timmers, H.T.M. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 2013, 33, 2067–2077.
- Li, K.; Wang, F.; Cao, W.-B.; Lv, X.-X.; Hua, F.; Cui, B.; Yu, J.-J.; Zhang, X.-W.; Shang, S.; Liu, S.-S.; et al. TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence. Cancer Cell 2017, 31, 697.e7–710.e7.
- Corcoran, C.A.; Luo, X.; He, Q.; Jiang, C.; Huang, Y.; Sheikh, M.S. Genotoxic and endoplasmic reticulum stresses differentially regulate TRB3 expression. Cancer Biol. Ther. 2005, 4, 1063–1067.
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339.
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400.
- Orea-Soufi, A.; Castillo-Lluva, S.; Salvador-Tormo, N.; Martín-Cabrera, P.; Recuero, S.; Gabicagogeascoa, E.; Moreno-valladares, M.; Mendiburu-Eliçabe, M.; Blanco-Gómez, A.; Ramos-Pittol, J.M.; et al. The Pseudokinase TRIB3 Negatively Regulates the HER2 Receptor Pathway and Is a Biomarker of Good Prognosis in Luminal Breast Cancer. Cancers 2021, 13, 5307.
- Flavin, R.; Peluso, S.; Nguyen, P.L.; Loda, M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010, 6, 551.
- Fhu, C.W.; Ali, A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 2020, 25, 3935.
- Prudente, S.; Hribal, M.L.; Flex, E.; Turchi, F.; Morini, E.; De Cosmo, S.; Bacci, S.; Tassi, V.; Cardellini, M.; Lauro, R.; et al. The functional Q84R polymorphism of mammalian Tribbles homolog TRB3 is associated with insulin resistance and related cardiovascular risk in Caucasians from Italy. Diabetes 2005, 54, 2807–2811.
More
