Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Vivi Li and Version 1 by Inês L. S. Delgado.
Monopolar spindle One Binder1 (MOB1) proteins regulate key cellular functions, namely cell multiplication and cell division. The unicellular parasite Toxoplasma gondii transitions between several morphological stages, with the need to control the number of parasites in its cellular environment. We hypothesized that MOB1 proteins could participate in the regulation of the T. gondii life cycle, having identified one MOB1 protein (TgMOB1) coded in its genome.
- Toxoplasma gondii
- tachyzoite
- MOB1
- Hippo pathway
- mitotic exit network
- apicomplexa
1. Introduction
Toxoplasma gondii is a unicellular parasitic eukaryotic organism that presents a highly polarized cell and an obligatory intracellular lifestyle during most of its life cycle. It is often considered the world’s most successful parasite as it is believed to infect all warm-blooded animals with frequently high prevalence rates. Part of the secret to its success is its ability to remain viable and mostly undetected in the host through regulatory mechanisms of proliferation and parasite number control. In the asexual life cycle phase, the parasites disseminate to different intermediate host tissues as rapidly proliferating tachyzoites that replicate by endodyogeny inside intracellular parasitophorous vacuoles [1]. Upon the onset of immunity, tachyzoites convert to slowly replicating bradyzoites that form tissue cysts, which may remain viable through the lifetime of the host [1]. Sexual reproduction of T. gondii takes place in the intestinal epithelium of felids, first going through schizogony driven asexual expansion, followed by gamogony, fecundation, and oocyst excretion into the environment, where two sporocysts containing eight sporozoites remain latent inside the very resistant oocyst wall [2]. Therefore, T. gondii presents a complex life cycle that relies on its ability to differentiate between several stages and regulate its proliferation.
Monopolar spindle One Binder (MOB) proteins form a conserved eukaryote family of kinase adaptors that regulate key cellular processes, namely chromosome segregation, mitotic exit, and cytokinesis [3,4,5,6,7,8][3][4][5][6][7][8]. MOB proteins also regulate cell polarity, contributing to the definition of cell morphology and accurate cell division, and are frequently present in structures that are fundamental for these processes, namely the spindle pole body and the centrosome [8,9,10,11][8][9][10][11]. The function of MOB proteins in cell division seems to be deeply conserved throughout the eukaryotic tree of life [4,5,7,8,12,13][4][5][7][8][12][13]. These proteins are pivotal members of the yeast mitotic exit network (MEN)/septation initiation network (SIN) and the multicellular eukaryote Hippo signaling pathway, where upstream signals lead to the activation of the core kinase module through a Cdc15p/MST1/2 kinase that in turn activates a Dbf2p/LATS1/2 kinase, enhanced by the MOB1 kinase adaptor [14,15,16,17][14][15][16][17]. MEN/SIN activation results in mitotic exit and cytokinesis through the effector proteins Cdc14p/Cip1p and Cdk1p, while Hippo signaling activation induces inhibition of pro-mitotic and anti-apoptotic associated transcription. Among the proteins that constitute the Hippo signaling pathway, MOB1 is the one that appears earlier in eukaryotes, supporting a core role for this protein [18]. Most eukaryotes, from budding yeast to humans, present more than one MOB protein [19]. This is also the case in Trypanosoma brucei—the only parasitic organism in which MOB1 proteins have been investigated to date—which possesses MOB1A and MOB1B proteins and in which MOB1 depletion leads to cytokinesis failure [5].
2. The Toxoplasma gondii genome encodes a single MOB1 protein homolog
To search for the presence of MOB genes in the parasite T. gondii, we performed a bioinformatics analysis by searching the ToxoDB.org database using as queries the deduced amino acid sequences of the best-characterized MOB1 proteins, the H. sapiens HsMOB1A and the S. cerevisiae ScMOB1. This search revealed the presence of one gene coding for a MOB1 protein, TgMOB1 (TGME49_304730). TgMOB1 is predicted to have 313 amino acids and a molecular mass of 34.9 kDa. This protein presents 24.5%/15.7% of identity and 38.9%/23.6% of similarity with the HsMOB1A and ScMOB1 sequences, respectively (Figure 1A). Additionally, we extended our search to other Hippo/MEN/SIN core members to assess the putative conservation of these pathways in T. gondii (Figure S1). We identified two NDR kinases that are probable homologs to Dbf/2/20p/LATS1/2 and NDR1/2 kinases, and an STE kinase that could perform a similar function to that of Cdc15p/MST1/2 kinases [49][20].
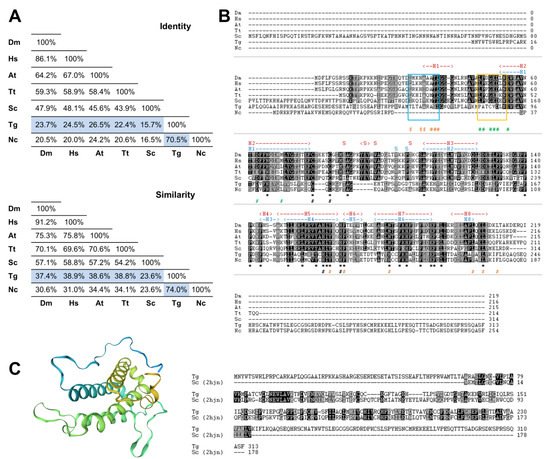

Figure 1. The Toxoplasma gondii MOB1 protein is highly conserved. The T. gondii MOB1 protein sequence was compared to other eukaryote MOB1 proteins. (A) Identity and similarity analysis of MOB1 proteins. The percentages of the T. gondii MOB1 versus other eukaryote MOB1 proteins assessed are highlighted in blue. (B) Alignment of MOB1 proteins. At the bottom of the alignment, asterisks (*) signal invariant residues. The conserved zinc-binding cysteine (T. gondii C105 and C107) and histidine residues (T. gondii H188 and H193) are indicated by a black # at the bottom. S. cerevisiae MOB1 residues that participate in the NDR–MOB1 interaction are indicated by a green # at the bottom [33][21]. The region of the NDR kinase binding motif LPXGED is highlighted by a yellow box [33][21]. ScMOB1 residues that participate in the MOB1 homodimer interaction are indicated by an orange # at the bottom [32][22]. The region of the ScMOB1 homodimer interaction-conserved residues integrating the helix H1 are highlighted by a light blue box [32][22]. The T. gondii MOB1 secondary structure, predicted by the SWISS-MODEL, is indicated at the top of the alignment in dark blue. The ScMOB1 secondary structure is indicated at the top of the alignment in dark red [32][22]. (C) T. gondii MOB1 theoretical 3D model based on the ScMOB179-314 structure (PDB h2jn) and corresponding sequence alignment. A rainbow color scheme from blue (N-terminus) to yellow (C-terminus) is presented. Dm, Drosophila melanogaster; Hs, Homo sapiens A; At, Arabidopsis thaliana A; Tt, Tetrahymena thermophila; Sc, Saccharomyces cerevisiae; Tg, Toxoplasma gondii; Nc, Neospora caninum.
Next, we compared the putative MOB1 sequence from T. gondii with MOB1 proteins from another eukaryote previously characterized. This comparison revealed that, as expected, TgMOB1 has the highest similarity with the MOB1 sequence from the related apicomplexan parasite Neospora caninum, and both diverge substantially in the N-terminal and C-terminal regions from the MOB1 sequences of the other eukaryotes assessed. Still, the central region of TgMOB1 contains the conserved residues corresponding to the MOB kinase activator family domain (T. gondii amino acids 69–229, PF03637) (Figure 1B). The zinc-binding cysteine and histidine pairs are conserved in all the species assessed. However, while the T. gondii and N. caninum MOB1 proteins present a CQC motif, the rest of the assessed species present a CXXXXC motif (Figure 1B). The ScMOB1 homodimer, detected in crystallography studies, is mediated by conserved residues that allow the association of a shallow hydrophobic depression and the amphipathic helix H1 (referred to as H0 in Mrkobrada et al. [32][22]). These residues are moderately conserved in T. gondii (Figure 1B). Of eight ScMOB1 residues identified as participating in the NDR–MOB1 interaction, the four residues located more in the C-terminal are conserved in T. gondii (D88, E91, D99, and L106) [33][21]. Of the more N-terminal residues, the NDR kinase binding motif LPXGED [33][21] is minimally conserved in T. gondii that presents a PATCVD motif instead (Figure 1B). To further analyze TgMOB1, we obtained a T. gondii MOB1 theoretical 3D structure model based on the known structure of the ScMOB179-314 region (PDB h2jn) (Figure 1C). The raw alignment included the residues 67–235 (54% coverage), 31% identity, a global model quality estimate (GMQE) of 0.33, and a qualitative model energy analysis distance constraint (QMEANDisCo) global score of 0.55 ± 0.07. The difference in identity score to that reported in Figure 1A occurs because the model alignment uses the ScMOB179-314, lacking the 78 amino acid N-terminal region that is specific to S. cerevisiae. The resulting model presents a core domain with an α helix-based structure organized in a bundle, common to MOB proteins and similar to the structure of the template ScMOB179-314. The results of the 3D model indicate high confidence in homology between proteins.
In order to investigate the evolutionary relationship between MOB1 proteins from different organisms, we performed a phylogenetic analysis, including parasites as well as model organisms (Figure 2). The first node separates animals from two other groups: (i) the group containing plants and unicellular organisms such as microalgae (Chlamydomonas reinhardtii) and ciliates (T. thermophila, Stentor coeruleus); and (ii) the group containing the remaining organisms. Focusing on the unicellular organisms included in the phylogenetic analysis, we observe that free-living and parasitic organisms are in different clades of the tree. Regarding parasitic organisms, a distinction between the Trypanosoma and Leishmania species, which belong to the supergroup Excavata, and parasites belonging to the phylum Apicomplexa (T. gondii, Hammondia hammondi, N. caninum, Eimeria tenella, Eimeria maxima, Cryptosporidium parvum, and Cryptosporidium muris) was detected. However, the MOB1 sequence from Giardia lamblia (Excavata) appears closer to the Apicomplexa than to the Trypanosomatida. The Apicomplexa form a monophyletic group in which cyst-forming coccidia (T. gondii, H. hammondi, N. caninum) appear very closely related and separated from the monoxenous genus Eimeria, while C. muris and C. parvum form a more distant group, mirroring the evolutionary history of these species. To date, no MOB proteins have been identified in hematozoan genomes, namely in Plasmodium spp., Babesia spp., or Theileria spp. [49][20].


Figure 2. Phylogenetic analysis of MOB1 proteins from different parasitic organisms. The predicted amino acid sequence of T. gondii MOB1 and those of different parasitic organisms were compared with MOB1 proteins from model organisms throughout the eukaryotic tree of life. The sequences were aligned and used to perform a maximum likelihood phylogenetic analysis. Kingdom (brown and green), phylum (blue), and ad hoc cluster names (black) are indicated on the right side. Bootstrap values are presented in red. The scale bar indicates the estimated number of amino acid substitutions per site.
References
- Black, M.W.; Boothroyd, J.C. Lytic Cycle of Toxoplasma Gondii. Microbiol. Mol. Biol. Rev. 2000, 64, 607–623.
- Tomasina, R.; Francia, M.E. The Structural and Molecular Underpinnings of Gametogenesis in Toxoplasma Gondii. Front. Cell. Infect. Microbiol. 2020, 10, 608291.
- Luca, F.C.; Winey, M. MOB1, an Essential Yeast Gene Required for Completion of Mitosis and Maintenance of Ploidy. Mol. Biol. Cell 1998, 9, 29–46.
- Luca, F.C.; Mody, M.; Kurischko, C.; Roof, D.M.; Giddings, T.H.; Winey, M. Saccharomyces Cerevisiae Mob1p Is Required for Cytokinesis and Mitotic Exit. Mol. Cell. Biol. 2001, 21, 6972–6983.
- Hammarton, T.C.; Lillico, S.G.; Welburn, S.C.; Mottram, J.C. Trypanosoma Brucei MOB1 Is Required for Accurate and Efficient Cytokinesis but Not for Exit from Mitosis. Mol. Microbiol. 2005, 56, 104–116.
- Galla, G.; Zenoni, S.; Marconi, G.; Marino, G.; Botton, A.; Pinosa, F.; Citterio, S.; Ruperti, B.; Palme, K.; Albertini, E.; et al. Sporophytic and Gametophytic Functions of the Cell Cycle-Associated Mob1 Gene in Arabidopsis Thaliana L. Gene 2011, 484, 1–12.
- Florindo, C.; Perdiga, J.; Fesquet, D.; Schiebel, E.; Pines, J.; Tavares, Á.A. Human Mob1 Proteins Are Required for Cytokinesis by Controlling Microtubule Stability. J. Cell Sci. 2012, 125, 3085–3090.
- Tavares, A.; Gonçalves, J.; Florindo, C.; Tavares, A.; Soares, H. Mob1: Defining Cell Polarity for Proper Cell Division. J. Cell Sci. 2012, 125, 516–527.
- Weiss, E.L.; Kurischko, C.; Zhang, C.; Shokat, K.M.; Drubin, D.G.; Luca, F.C. The Saccharomyces Cerevisiae Mob2p—Cbk1p Kinase Complex Promotes Polarized Growth and Acts with the Mitotic Exit Network to Facilitate Daughter Cell—Specific Localization of Ace2p Transcription Factor. J. Cell Biol. 2002, 158, 885–900.
- Chalker, U.L.; Frankel, J. Morphogenesis: A Mob Rules from the Rear. Curr. Biol. 2014, 24, R700–R702.
- Bae, J.S.; Kim, S.M.; Jeon, Y.; Sim, J.; Jang, J.Y.; Son, J.; Hong, W.; Park, M.K.; Lee, H. Loss of Mob1a/b Impairs the Differentiation of Mouse Embryonic Stem Cells into the Three Germ Layer Lineages. Exp. Mol. Med. 2019, 51.
- Lai, Z.-C.; Wei, X.; Shimizu, T.; Ramos, E.; Rohrbaugh, M.; Nikolaidis, N.; Ho, L.-L.; Li, Y. Control of Cell Proliferation and Apoptosis by Mob as Tumor Suppressor, Mats. Cell 2005, 120, 675–685.
- Wilmeth, L.J.; Shrestha, S.; Montaño, G.; Rashe, J.; Shuster, C.B. Mutual Dependence of Mob1 and the Chromosomal Passenger Complex for Localization during Mitosis. Mol. Biol. Cell 2010, 21, 380–392.
- Bardin, A.J.; Amon, A.; Medical, H.H. Men and sin: what’s the difference? Nature 2001, 2, 815–826.
- Hotz, M.; Barral, Y. The Mitotic Exit Network: New Turns on Old Pathways. Trends Cell Biol. 2014, 24, 145–152.
- Hergovich, A. Hippo Signaling in Mitosis: An Updated View in Light of the MEN Pathway; Monje-Casas, F., Queralt, E., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1505, ISBN 9781493965021.
- Duhart, J.C.; Raftery, L.A. Mob Family Proteins: Regulatory Partners in Hippo and Hippo-Like Intracellular Signaling Pathways. Front. Cell Dev. Biol. 2020, 8, 1–22.
- Sebé-Pedrós, A.; Zheng, Y.; Ruiz-Trillo, I.; Pan, D. Premetazoan Origin of the Hippo Signaling Pathway. Cell Rep. 2012, 1, 13–20.
- Hergovich, A. MOB Control: Reviewing a Conserved Family of Kinase Regulators. Cell. Signal. 2011, 23, 1433–1440.
- Delgado, I.L.S.; Carmona, B.; Nolasco, S.; Santos, D.; Leitão, A.; Soares, H. Mob: Pivotal Conserved Proteins in Cytokinesis, Cell Architecture and Tissue Homeostasis. Biology 2020, 9, 413.
- Parker, B.W.; Gogl, G.; Bálint, M.; Hetényi, C.; Reményi, A.; Weiss, E.L. Ndr/Lats Kinases Bind Specific Mob-Family Coactivators through a Conserved and Modular Interface. Biochemistry 2020, 59, 1688–1700.
- Mrkobrada, S.; Boucher, L.; Ceccarelli, D.F.J.; Tyers, M.; Sicheri, F. Structural and Functional Analysis of Saccharomyces Cerevisiae Mob1. J. Mol. Biol. 2006, 362, 430–440.
More
