The T. delbrueckii Kbarr2 killer strain contains two M killer viruses (Mbarr1 and M1) and a LBC virus (TdV-LBCbarr2), which has helper capability to maintain both M viruses. The genomes of TdV-LBCbarr1 and TdV-LBCbarr2 were characterized by high-throughput sequencing (HTS). Both RNA genomes share sequence identity and similar organization with their ScV-LBC counterparts. They contain all conserved motifs required for translation, packaging, and replication of viral RNA. Their Gag-Pol amino-acid sequences also contain the features required for cap-snatching and RNA polymerase activity. However, some of these motifs and features are similar to those of LA viruses, which may explain that at least TdV-LBCbarr2 has a helper ability to maintain M killer viruses.
1. Analysis of the dsRNA Genomes from TdV-LBCbarr1 and TdV-LBCbarr2
Two different sequences were obtained from the L dsRNA (agarose electrophoresis band of 4.6 kb) present in the Kbarr1 EX1180 strain, one similar to that of LA virus genomes (named TdV-LAbarr1) that was previously characterized
[1][12], and another sequence similar to that of LBC viruses (named TdV-LBCbarr1). However, only one sequence was obtained from the L dsRNA present in Kbarr2 EX1257 strain (named TdV-LBCbarr2). This strain contains two killer viruses, Mbarr1 and M1
[2][3], which indicates that TdV-LBCbarr2 is the helper virus required for replication and maintenance of both M viruses.
The complete sequence found for TdV-LBCbarr1 cDNA was 4763 nucleotides in length, and 5115 nt for TdV-LBCbarr2, which are slightly larger than the size estimated by agarose-gel electrophoresis (
Table 1). Most of the sequence of both LBCbarr genomes (4565 nt central stretch) showed about 52% nucleotide identity with the previously known ScV-LBC1-original and ScV-LBClus4 genomes (
Figure 1,
Figures S1 and S3), while it only shared 39% identity with
Saccharomyces and
Torulaspora LA genomes. It was considered this central stretch as the canonical sequence of LBCbarr genomes, and sequences upstream and downstream as 5′- and 3′-extra sequences, respectively; as it was previously considered for V-LA and V-M genomes
[2][1][3,12]. The canonical sequences of TdV-LBCbarr1 and TdV-LBCbarr2 genomes share great identity (94%), which means that both are the same virus with some nt-sequence differences. Therefore,
it w
ase mainly focused on the TdV-LBCbarr2 sequence for genome comparison with other L viruses.
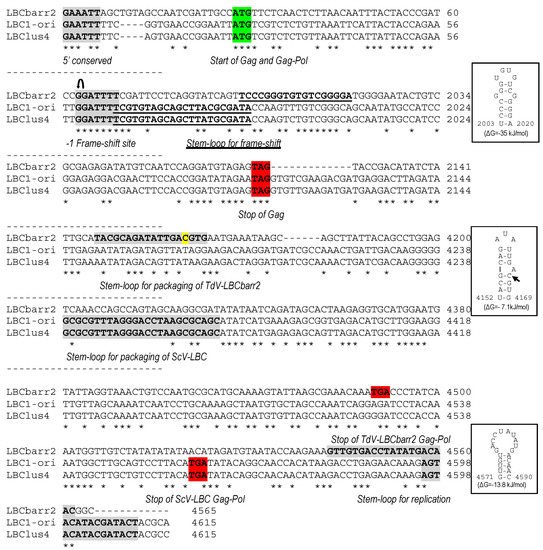
Figure 1. Partial multiple sequence alignment between ScV-LBC1-original, ScV-LBClus4, and TdV-LBCbarr2 (+) strand nucleotide sequences (cDNA). The full sequence alignment is presented in
Supplemental Material (Figure S1). 5′GAA(A/T)TT conserved motif (5′ conserved), translation initiation (start of Gag and Gag-Pol), termination (stop of Gag or Gag-Pol) codons, ribosome frameshifting site (−1 frameshift site), frameshifting associated sequence (stem-loop for frameshift), packaging signal (stem-loop for packaging), and replication signal (stem-loop for replication) are indicated, shaded and/or underlined in the nucleotide sequence. Different residue in LBCbarr2 with respect to LBCbarr1 virus is yellow shaded.
, ribosomal frameshift. Asterisks (*) indicate identical nucleotide positions. The secondary structures of the putative cis signals for frameshifting, packaging, and replication of TdV-LBCbarr2 are displayed at the right of the sequence panel.
Table 1. Characteristics of the dsRNA LBC-virus genomes sequenced by HTS.
| Virus |
Previous Sequenced Length (bp)/Yeast Strain |
Newly Analysed Yeast Strain |
Killer Phenotype/dsRNA Isotype |
Sequenced Length (bp) (Canonical) |
| LBC1 |
4615/ | Sc | 299 (LBC1-original) |
Sc | EX231 |
K1/M1-1 |
4971 (4615) |
| LBC2 |
4614/ | Sc | S3920 |
Sc | EX1125 |
K2/M2-4 |
4722 (4615) |
| LBClus |
4614/ | Sc | EX229 |
Sc | EX229 |
Klus/Mlus4 |
4839 (4615) |
| Sc | EX1160 |
Klus/MlusA |
4661 (4615) |
| LBCbarr-1 |
Unknown/ | Td | EX1180 |
Td | EX1180 |
Kbarr1/LBCbarr1 |
4763 (4565) |
| LBCbarr-2 |
Unknown/ | Td | EX1257 |
Td | EX1257 |
Kbarr2/LBCbarr2 |
5115 (4565) |
Sc, Saccharomyces cerevisiae. Td, Torulaspora delbrueckii.
2. Analysis of the Gag-Pol Sequences from TdV-LBCbarr1 and TdV-LBCbarr2
No relevant identity was found between the putative Gag-Pol amino acid sequences of both TdV-LBCbarr and those of LA viruses (24–24.8%), similarly to that found between Gag-Pol of ScV-LBC1-original and ScV-LA1-original (24.6%). TdV-LBCbarr Gag-Pol sequences showed 44% global identity with that of ScV-LBC1-original (
Figure 23, Figure
45 and
Figure S2). Despite this modest identity, both LBCbarr Gag-Pol sequences contain most of the key motifs for Gag and Pol functions previously found in
Saccharomyces LBC viruses. This identity is much lower than that found among Gag-Pol sequences of known
S. cerevisiae LBC-viruses, which is greater than 95% (
[3][4] and see below). This modest identity increases to 64–66% when comparing only the highly-conserved RdRp-domain of TdV-LBCbarr and ScV-LBC polymerases. This domain contains the five motifs (3, A, B, C, and D) conserved among RdRps, which are essential for RNA polymerase activity
[4][5][23,24]. The most conserved amino acids of these five motifs are identical in all RdRps of yeast L viruses except one amino acid in motif A, which is “DYDDFNS” in all known
Saccharomyces and
T. delbrueckii LA viruses, and “DFDDFNS” in all
S. cerevisiae LBC viruses. Surprisingly, motif A of both TdV-LBCbarr polymerases is identical to that found in LA viruses. However, surrounding residues of these five conserved amino acids of LBCbarr polymerases are in most cases coincident with those of
S. cerevisiae LBC viruses (
Figure 34).
Figure 23. Comparison between partial amino-acid sequences of Gag-Pol encoded by ScV-LBC1-original, ScV-LBClus4, and TdV-LBCbarr2 genomes. The full sequence alignment is presented in
Supplemental Material (Figure S2). The separation between Gag and Pol is indicated (Gag◄►Pol). The H156 residue required for 5′cap-snatching is black shaded. The four crucial residues for cap recognition (Tyr-152, Asn-154, Tyr-452 and Trp-545) are grey shaded. The highly conserved central third of Pol (RdRp domain) is underlined, the five consensus motifs (3, A, B, C, and D) conserved in RNA-dependent RNA polymerases from Totiviruses are indicated above the sequence, and the conserved amino acids for each motif are grey shaded. Different residue in TdV-LBCbarr with respect to ScV-LBC is yellow shaded. Asterisks (*) indicate identical amino acids; colons (:) and single dots (.) indicate conserved and semi-conserved amino acids, respectively.
Figure 34. Conserved motifs in the Gag-Pol RdRp domain of L viruses from S. cerevisiae and T. delbrueclii. The extent of each domain is indicated in the top of the figure. The last amino acid of the stretch containing each motif is indicated to the right of the sequence. Asterisks (*) indicate identical amino acids; colons (:) and single dots (.) indicate conserved and semi-conserved amino acids, respectively. Conserved identical residues in all virus motifs are grey shaded. Different residues in LBC with respect to LA viruses are yellow shaded. Different residues for TdV-LBCbarr and ScV-LBC1-original are pink shaded. “Y” residue in motif A of TdV-LBCbarr that is coincident with LA viruses is indicated inside a square.
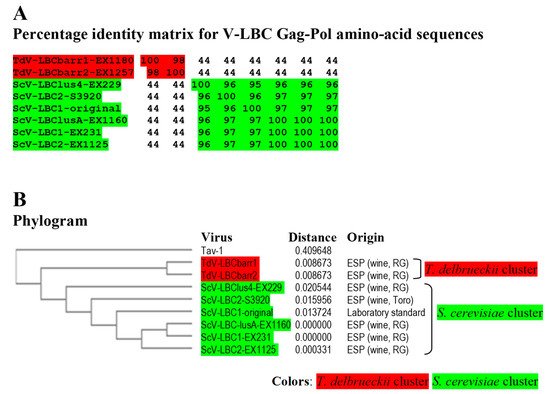
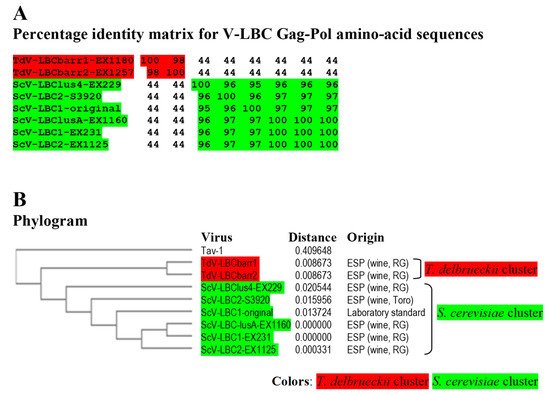
Figure 4. Phylogenetic relationship of yeast LBC viruses. (A) Percentage identity matrix between the complete amino acid sequences of the Gag-Pol proteins of LBC viruses. Each identity value was rounded to the nearest whole number. (B) Phylogram with evolutionary distances (given by the MUSCLE program) and geographical location at which each killer yeast strain was isolated. RG, Ribera del Guadiana region in Extremadura. ESP, Spain. Viruses included in each of the two main clusters, as well as their identity values, are shaded in red or green. The Tuber aestivum (Tav-1) totivirus was used as outgroup (GenBank accession number HQ158596.1).
The identity between Gag sequences of TdV-LBCbarr and those of ScV-LBC viruses also is low, about 24–37.8%. However, the Gag His154 residue of ScV-LA1-original (that is His155 in ScV-LBC1-original, and His156 in TdV-LBCbarr) required for the cap-snatching mechanism in the virion
[6][25], and the four putative crucial residues for cap recognition (Tyr-150, Asp-152, Tyr-452, and Tyr-538 in ScV-LA1-original; that are Tyr-152, Asp-154, Tyr-449 and Trp-543 in ScV-LBC1-original
[7][26]) are also present in the Gag protein of both LBCbarr viruses (Tyr-152, Asn-154, Tyr-452 and Trp-545;
Figure 23 and
Figure S2). In addition to the slightly different location for each residue in the Gag sequence, the main difference among these viruses is that the forth residue is Trp in LBC1-original and LBCbarr viruses, while it is Tyr in ScV-LA1-original. The same difference was found when comparing all known LA and LBC viruses from
Saccharomyces and
T. delbrueckii yeasts (data not shown).
3. Comparison of TdV-LBCbarr with LBC Viruses from Saccharomyces Yeasts
The dsRNA and Gag-Pol sequences of both TdV-LBCbarr were compared with their counterparts from
S. cerevisiae to analyze their phylogenetic relationship. The genomic sequence of ScV-LBC1-original, ScV-LBClus-EX229, and ScV-LBC2-S3920 were already known (
Table 1 and
Table 2). The genomes of ScV-LBC1 from strain EX231, ScV-LBC2 from EX1125, and ScV-LAlusA from EX1160 were
de novo sequenced by HTS techniques for this study. ScV-LBClus4 from EX229, previously sequenced by Sanger procedure and named ScV-L-BC-lus
[3][4], was also de novo sequenced to assess the accuracy of HTS procedure. Sequences obtained by both procedures share 99.9% identity. Only six nucleotide changes were found in the genome of ScV-LBClus4 with respect to ScV-L-BC-lus: C548A, T704C, T2667G, G2668C, G2669T, and G2671A. As nucleotides of ScV-LBClus4 (HTS) in these positions coincided best with the rest of the
S. cerevisiae LBC viruses, this sequence was the one used for further comparison.
Table 2. Previously known nucleotide sequence of LBC and LA viruses used in this study.
| Virus |
Accession Number |
Reference/Comment |
| ScV-LBC1-original |
U01060.1 |
Previously [8] known as ScV-La or L-B-C. Renamed in this study to distinguish it from other LBC viruses from different K1 killer yeasts | Previously [7] known as ScV-La or L-B-C. Renamed in this study to distinguish it from other LBC viruses from different K1 killer yeasts |
| ScV-LBClus4-EX229 |
KT784813.1 |
[3] | [4] |
| ScV-LBC2-S3920 |
KX906605.1 |
[3] | [4] |
| TdV-LAbarr1-EX1180 |
MW174763 |
[1] | [12] |
| ScV-LA1-original |
J04692.1 |
Previously [9] known as ScV-LA. Renamed in this study to distinguish it from other LA viruses from different K1 killer yeast strains | Previously [11] known as ScV-LA. Renamed in this study to distinguish it from other LA viruses from different K1 killer yeast strains |
| ScV-LA1-EX231 |
MW174760 |
[1] | [12] |
| ScV-LAlus-EX229 |
JN819511.1 |
[10] | [13] |
| ScV-LAlus4-EX229 |
MW174758 |
[1] | [12] |
| ScV-LAlus1-EX436 |
MW174761 |
[1] | [12] |
| ScV-LAlusA-EX1160 |
MW174762 |
[1] | [12] |
| ScV-LA2-S3920 |
KC677754.1 |
[3] | [4] |
| ScV-LA2-EX1125 |
MW174759 |
[1] | [12] |
| SpV-LA28 |
KU845301.2 |
Formerly [11] assigned to | Formerly [27] assigned to | S. cerevisiae | but recently re-assigned to | S. paradoxus [12] | [6] |
Sc, Saccharomyces cerevisiae. Td, Torulaspora delbrueckii. Sp, Saccharomyces paradoxus.
Identity among Sc-LBC viruses was similar to that previously found
[3][4], above 95% for amino acid sequences. Two clusters were found to include all LBC viruses: S. cerevisiae cluster, in which ScV-LBClusA-EX1160, ScV-LBC1-EX231 and ScV-LBC2-EX1125 isolated from Extremadura seem to be the same virus (99.9–100% identity of Gag-Pol); and T. delbrueckii cluster grouping TdV-LBCbarr1-EX1180 and TdV-LBCbarr2-EX1257, that were also isolated from Extremadura, and also appear to be the same virus (98.3%). Viruses from different clusters share a fairly low identity rate (43.5–44.2%), despite that most viruses were isolated from the same region. Given the high similarity of ScV-LBClus4-EX229, ScV-LBC2-S3920 and ScV-LBC1-original with the rest Sc-LBC virus (95–100%), they appear to be just variants of the same virus (
Figure 45). It is noteworthy that the same LBC virus was found can coexist with different killer M viruses in different yeast strains (
Table 1).
Figure 5. Phylogenetic relationship of yeast LBC viruses. (A) Percentage identity matrix between the complete amino acid sequences of the Gag-Pol proteins of LBC viruses. Each identity value was rounded to the nearest whole number. (B) Phylogram with evolutionary distances (given by the MUSCLE program) and geographical location at which each killer yeast strain was isolated. RG, Ribera del Guadiana region in Extremadura. ESP, Spain. Viruses included in each of the two main clusters, as well as their identity values, are shaded in red or green. The Tuber aestivum (Tav-1) totivirus was used as outgroup (GenBank accession number HQ158596.1).
4. Analysis of TdV-LBCbarr Genomes
The average nucleotide identity between both T. delbrueckii LBCbarr viruses and S. cerevisiae LBC viruses (52%) was lower than that found between LA viruses of the same yeast species (60%)
[1][12]. Moreover, this nucleotide identity was much lower than that found between Sc-LBC viruses (89–100%). This can be expected because these two yeast species are usually found in different ecological niches
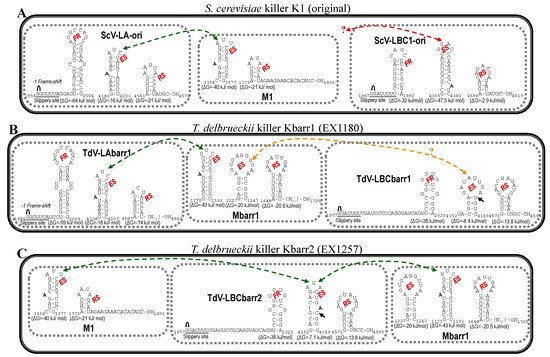
[1][13][14][12,28,29]. This finding raises the possibility that LBCbarr viruses are a new type of virus different from the already known LA and LBC viruses. Notwithstanding this, the organization of both TdV-LBCbarr genomes is similar to that of ScV-LA and ScV-LBC. They contain the same two ORFs, Gag and Gag-Pol. However, while TdV-LA and ScV-LA share 87.5–100% identity in some regions considered important for the virus replication (such as the frameshifting region required for translation of Gag-Pol fusion protein, or the virus packaging signal), TdV-LBC and ScV-LBC only share the ribosome slippery site located upstream of the Gag ORF stop codon (GGAUUUU). Interestingly, the three stem-loops involved in frameshifting, RNA binding and packaging, and RNA replication of TdV-LBCbarr genomes resemble those secondary structures found in similar positions in ScV-LA genomes, especially those of TdV-LBCbarr2 (
Figure 52). This may explain why Sc-LA and Td-LBCbarr viruses have capability to maintain killer M viruses; whereas Sc-LBC viruses that have these stem-loops located in other positions do not have not this capability. TdV-LBCbarr2 contains a stem-loop for packaging and encapsidation (ES signal) similar to the ES of S. cerevisiae LA and M1 viruses, and also similar to the ES of TdV-Mbarr1. This may explain why TdV-LBCbarr2 has capability to maintain both M1 and Mbarr1 viruses in the same cell. However, a difference of only one nucleotide in LBCbarr1 ES with respect to LBCbarr2 ES (C4166T) changes its secondary structure, making LBCbarr1 probably unable to maintain the M1 virus. Optionally, the ES secondary structure of LBCbarr1 is similar to a newly identified possible ES in the Mbarr1 genome, raising the possibility that LBCbarr1 may maintain Mbarr1 at the same time that TdV-LAbarr1 does (
Figure 52). I.e., Mbarr1 could be maintained by two different helper viruses in EX1180 strain, Td-LAbarr1 and Td-LBCbarr1.
It was confirmed the presence of all motifs and features previously proposed as required for viral replication of ScV-LBC1-original in all the newly sequenced S. cerevisiae LBC genomes; and the particular location of the stem-loop proposed for frameshifting, that overlaps 1 nt with the ribosome slippery site
[3][4] (
Figure 52). The ribosome frame shift in L viruses happens with low frequency, in about 1% of ribosomes
[15][30]. The overlapping of the slippery site and the frameshifting stem-loop may imply a steric effect that decreases the frame shift frequency, reducing the amount of Cap-Pol required for viral replication. This may explain, at least in part, the low relative amount of LBC virus with respect to LA virus in S. cerevisiae
[16][17][9,31]. In addition to this, the genome location and secondary structure of the stem-loops involved in viral replication, that is different to those of Sc-LA and Td-LBCbarr viruses, may account for the inability of Sc-LBC viruses to behave as helper virus to maintain killer M viruses.
Figure 5. Secondary structure representations of signals for frameshifting, encapsidation, and replication present in the (+)RNA strand of L and M viruses from killer yeasts: (A) S. cerevisiae K1 (original), (B) T. delbrueckii Kbarr1 (EX1180), and (C) T. delbrueckii Kbarr2 (EX1257). FR, frameshifting region. ES, encapsidation signal. RS, replication signal.  , ribosomal frameshift. Black arrow indicates the single nucleotide changes between ES of LBCbarr1 and LBCbarr2. Green dot-line arrow indicates equivalent ES for L and M viruses in the same yeast. Orange dot-line arrow indicates possible equivalent ES for LBCbarr1 and Mbarr1 in Kbarr1 EX1180 yeast. Red dot-line arrow indicates unknown equivalent ES in M viruses for the ES of ScV-LBC1-original.
, ribosomal frameshift. Black arrow indicates the single nucleotide changes between ES of LBCbarr1 and LBCbarr2. Green dot-line arrow indicates equivalent ES for L and M viruses in the same yeast. Orange dot-line arrow indicates possible equivalent ES for LBCbarr1 and Mbarr1 in Kbarr1 EX1180 yeast. Red dot-line arrow indicates unknown equivalent ES in M viruses for the ES of ScV-LBC1-original.
 , ribosomal frameshift. Asterisks (*) indicate identical nucleotide positions. The secondary structures of the putative cis signals for frameshifting, packaging, and replication of TdV-LBCbarr2 are displayed at the right of the sequence panel.
, ribosomal frameshift. Asterisks (*) indicate identical nucleotide positions. The secondary structures of the putative cis signals for frameshifting, packaging, and replication of TdV-LBCbarr2 are displayed at the right of the sequence panel.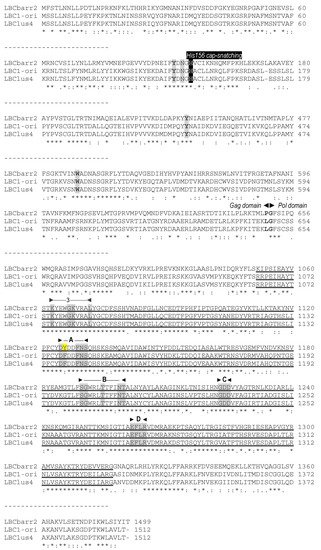

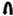 , ribosomal frameshift. Black arrow indicates the single nucleotide changes between ES of LBCbarr1 and LBCbarr2. Green dot-line arrow indicates equivalent ES for L and M viruses in the same yeast. Orange dot-line arrow indicates possible equivalent ES for LBCbarr1 and Mbarr1 in Kbarr1 EX1180 yeast. Red dot-line arrow indicates unknown equivalent ES in M viruses for the ES of ScV-LBC1-original.
, ribosomal frameshift. Black arrow indicates the single nucleotide changes between ES of LBCbarr1 and LBCbarr2. Green dot-line arrow indicates equivalent ES for L and M viruses in the same yeast. Orange dot-line arrow indicates possible equivalent ES for LBCbarr1 and Mbarr1 in Kbarr1 EX1180 yeast. Red dot-line arrow indicates unknown equivalent ES in M viruses for the ES of ScV-LBC1-original.