You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Dean Liu and Version 1 by Rwik Sen.
A review on COVID-19 in the context of developmental biology, exosomes, and transcriptome.
- SARS-CoV-2
- COVID-19
- placental transmission
- development
- exosomes
- transcriptome
[1]1. SARS-CoV-2 and Fetal Development: The Role of Exosomes
Although it is clear that SARS-CoV-2 infection induces an immune response in pregnant women, the alterations in the fetal immune responses remain a matter of intense debate. 205 infants born to COVID-19-positive mothers were investigated [59][1]. While only ~10% infants were found to be positive for COVID-19, most studied infant cases had developed immunoglobulin G and M (IgG, IgM) antibodies against SARS-CoV-2 [59][1]. In another study, no viral RNA was detected in the placentas of COVID-19 positive pregnant women [60][2]. Furthermore, there seems to be no confirmed cases of intrauterine infection of SARS-CoV-2 from mothers to their fetuses. Although severe illness has been seen in infants younger than 1 year, such cases have had confirmed underlying comorbidities [61][3]. These findings suggest that vertical infection is rare, and a natural passive immunity is developed in infants who are born to mothers with COVID-19 [59,60][1][2].
Exosomes are secreted by all cell types studied to date [62,63,64][4][5][6]. With respect to the placental lineage, exosomes have been investigated from mesenchymal, endothelial, and trophoblastic lineages and have been demonstrated to suppress T-cell expression [65][7]. Here, researchers investigated the role of exosome trafficking in utero and their significance with respect to SARS-CoV-2 infections and the subsequent development of an immune response in infants. Exosomes are extracellular nanovesicles (~50–200 nm) of endocytic origin that package cellular constituents; this is likely to maintain cellular homeostasis, but the reason behind their production is unknown [66][8]. There seem to be two potential hypotheses of exosomal contribution in utero, as well as in fetal development: (1) SARS-CoV-2 infections via exosomes, or (2) in utero development of immunity. While exosomes have been found to carry viral RNA, there seems to be little to no viral replication in utero [60][2]. This observation discards the first hypothesis that exosomes may induce viral infection in utero.
Alternatively, our second hypothesis regarding exosomes’ role in the development of an immune response is of major interest, and may have numerous implications in utero and in fetal development [67,68][9][10]. Exosomes predominantly carry major histocompatibility complex (MHC) class I and II molecules on their surface, which can activate T lymphocytes, and trigger an adaptive immune response [67][9]. As the placenta promotes the production of exosomes that are enriched in developmental and immune response cargo, the presence of a complete antigen-presentation molecular machinery within exosomes has direct implications on the development of a fetal immune response to SARS-CoV-2 infection [65][7]. Rising debate surrounding whether exosomes are capable of either a direct or indirect activation of immune response may be correlated to the cell of origin of the given exosomes.
Regardless, accumulating evidence suggests that MHC-I and II molecules on the surface of exosomes can functionally form a complex with antigenic peptides to induce immune activation. The presence of these MHC-I and II molecules may suggest and provide the basis of the important roles of exosomes in the immune cascade. Based on these observations, we reason that the mechanism of MHC secretion through exosomes and their potential role in cell-to-cell communication, targeted function, and immune regulation may target in utero immune development. Finally, we hypothesized the potential use of MHC-exosomes as the extracellular particles of choice, which may lead to therapeutic procedures for immunity development in mothers and infants.
2. Development and Functions of Exosomes: Biogenesis and Biology
Exosomes are a subclass of extracellular vesicle (Evs) that are released by all cell types and are involved in extracellular communication. Unlike other Evs, exosomes are formed by the inward budding of the membrane of late endosomes, otherwise known as multivesicular bodies (MVBs) [69,70,71][11][12][13]. Subsequently, these MVBs fuse with the plasma membrane (PM) resulting in the release of exosomes to the extracellular environment [72,73][14][15]. Given their unique intracellular trafficking pathway, exosomes encapsulate different cargo content [74,75,76,77][16][17][18][19]. The endosomal sorting complexes required for transport (ESCRT) proteins, along with the Rab (Ras-associated binding) small GTPase family, serve a crucial role in the modulation of exosomal secretion and trafficking [78][20].
This mechanism starts with the ESCRT-0 protein utilizing hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) to identify and cluster ubiquitinated transmembrane proteins in the endosomal membrane. Once properly localized, the HRS recruits ESCRT-I/II complexes, along with associated proteins (for instance, TSG101, ALIX, VPS4, etc.), for the initiation of MVB biogenesis via budding. Finally, the actual process, involving vesicle scission, is primarily driven by the ESCRT-III protein. Free ESCRT components and ubiquitin molecules are recycled for repeating the process post-scission of the MVBs [74,78][16][20].
Following the formation of MVBs, the remainder of the trafficking pathways (comprising the cytoskeleton, molecular motors, and vesicle fusion machinery) are mostly regulated by the Rab family of small GTPases [79][21]. In particular, both RAB27A and RAB27B are associated with the promotion of MVB docking and fusing to the PM, as well as the vesicle transfer from the Golgi apparatus to MVBs. Likewise, mechanisms involving RAB small GTPases often recruit SNAP receptors (SNAREs), a superfamily of proteins, for the mediation of vesicle trafficking within cells [79,80][21][22].
Despite the critical role of the ESCRT complexes, further evidence has demonstrated an alternative, ESCRT-independent, pathway of exosomal packaging and formation [80][22]. In addition to proteins that are actively involved in exosomal biogenesis (i.e., TSG101, ALIX, RAB proteins, and annexins), other frequently observed exosomal proteins include membrane transport proteins, metabolic enzymes, fusogenic proteins, tetraspanins, heat shock proteins, cytoskeletal proteins (actin and tubulin), lipoproteins, and enzymes (phospholipases).
Nevertheless, exosomes are not the only extracellular vesicles that are released; others which are released are often called apoptotic, micro-, and onco-vesicles [69,71][11][13]. All extracellular vesicle cargo leave molecular footprints from their cell of origin; exosomes selectively package proteins and nucleic acids and appear to avoid cellular debris [69,81][11][23]. Recent proteomic studies have revealed a set of endocytic, cytoplasmic, and endosomal proteins in exosomes. In contrast, PM-derived vesicles predominantly contain nuclear DNA, mitochondrial DNA, rRNA, and PM-associated proteins [69,81][11][23].
3. Genomic, Transcriptomic, Proteomic, and Lipidomic Landscape of Exosomes
Despite the wide therapeutical and diagnostic applications that have been confirmed today, exosomes were thought to only pertain to cargo cell debris and waste in the early stages of exosomal research in the 1980s [78,82][20][24]. Nevertheless, beginning in the 1990s, studies showed results suggesting exosomes’ pivotal role in cell-to-cell communication and as triggers for cancer immune responses [83,84][25][26]. The proteomic composition of exosomes is a consequence of their cell of origin and their endosomal molecular pathways. Exosomes are composed of highly enriched tetraspanins (CD9, CD81, CD63) and other endosome-associated proteins (RAB, SNARE, TSG101, ALIX, and ESCRT) [64,70,71][6][12][13]. Much of the proteomic landscape of exosomes has been discussed above in the exosome’s biogenesis section.
Major breakthroughs were marked in 2000′s, as mRNAs and microRNAs were unveiled in exosomes along with their influence on cellular behaviors and functions [81][23]. In particular, a wide variety of genetic material were gradually identified, including mRNA, ncRNA, miRNA, lncRNA, ssDNA, dsDNA, mitochondrial DNA, and oncogene amplifications [81,85,86,87,88][23][27][28][29][30]. In chronic lymphocytic leukemia, exosomes shuttle proteins, lipids, miRNAs and mRNAs to recipient cells, and regulate their transcriptomes and behaviors [89][31]. Those exosomes are enriched for miR-202-3p, which likely impacts Hedgehog signaling [89][31]. Exosomes contain mRNA and a variety of non-coding RNA whose alterations are partly reflected in the cellular transcriptome, which indicates the potential of the exosomal transcriptome as a biomarker [90,91][32][33]. Exosomes that are derived from the placenta regulate maternal immune tolerance during pregnancy [92][34]. The transcriptomes and proteomes of exosomes that are derived from avian serum have provided important insights regarding antiviral vaccination [93][35]. Hence, exosomal transcriptome analysis has the potential to inform about some of the complexities that have been reported in pregnant women who have received COVID-19 vaccines [94][36]. Exosomes are also significant for other diseases, because their participation in signaling pathways and cell-to-cell communication impacts the tumor microenvironment [62,63,91,95][4][5][33][37].
Aside from genetic materials, exosomes have also been confirmed to deliver lipids and proteins [74,76][16][18]. The inclusion of proteomic components and genetic materials suggests that exosomes have the capability of regulating and triggering specified signaling cascades, thereby altering the transcriptional landscape of the targeted cell. These characteristics enable exosomes to regulate cellular crosstalk and vesicle trafficking for impacting disease progression, the tumor microenvironment, metastasis, and other processes [76][18].
When compared with their cell or origin, exosomes tend to be enriched in proteins that are located in lipid rafts, including glycosylphosphatidylinositol-anchored proteins and flotillin. Exosomes are enriched in cholesterol, sphingomyelin, gangliosides, ceramides, and phosphatidylserine (PS) [64][6]. PS and phosphatidylethanolamine (PE) are enriched on the outer membrane of exosomes, whereas PS and PE tend to be depleted in the outer cell membrane. The underlying reason behind the differential lipid composition of exosomes could be credited, in part, to a different membrane curvature from their cell of origin. The large curvature of cells (~2 micron) versus the small curvature of exosomes (~100 nm) may recruit different lipids for their formation.
4. Entry of SARS-CoV-2 and Exosomes in Cells
Most cells internalize ligands through multiple mechanisms, such as phagocytosis or pinocytosis [98,99][38][39]. Additionally, membranous particles can also fuse with the cellular PM, however, this mechanism is less profound in the internalization of influenza viruses [98][38]. In fact, most viruses are believed to follow the endosomal pathways and avoid membrane-membrane fusion with the cells [98][38]. This may be because membrane fusion can assimilate viral proteins and lipids with the PM of the cells and leave traces at the cell surface. These leftover traces may help cells to recognize the incoming intruders and be better prepared for the next similar viral invasion.
With respect to the SARS-CoV-2 virus, it is now established that CoVs use the ACE2 receptor for binding with the host cells; gastrointestinal, kidney, and heart tissues express the highest amount of ACE2 [2][40]. Serine protease TMPRSS2 acts as the priming agent for the S protein [2,96][40][41]. It has also been confirmed that the spike protein facilitates viral entry into the host cell [96,97][41][42]. Taken together, the most likely mechanism of SARS-CoV-2 entry seems to be a receptor-mediated endocytosis or pinocytosis. Whether the entry is clathrin-dependent or independent remains to studied.
5. Exit of SARS-CoV-2 and Exosomes from Cells
Recent studies have shown that CoVs use lysosomal, instead of biosynthetic pathways, to exit the cells [28][43]. Ideally, a late endosome (500–1000 nm) encapsulates the CoV in the cytoplasm of the hijacked cells. Subsequently, the endosome fuses with the PM to release the virus particles to the extracellular environment. This mechanism is identical to the biogenesis and release of exosomes [66,69][8][11].
Here we propose a hypothesis (Figure 3): viruses hijack the evolutionary exosomal pathways, and current vaccination strategies could learn from exosomal intake and release for a better understanding of viral entry, intake, and release. Studies on cancer have shown that exosomes and exosomal RNA impact cellular development, function, and gene expression [100,101,102,103][44][45][46][47]. Hence, the next section describes the impact of SARS-CoV-2 infection upon the host transcriptome, readouts from host transcriptomic perturbations, and lessons from the viral transcriptome which have been obtained from infected samples of COVID-19 patients.
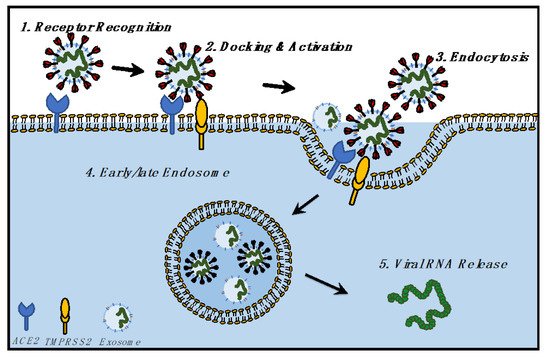

Figure 3. Hypothesis of the biological relationship between SARS-CoV-2 and exosomes, focusing on the identical mechanism followed by SARS-CoV-2 and exosomes for cellular entry and exit.
Discussion on the developmental biology and transcriptome aspects of COVID-19 can be found in Reference 1. All content and citations included in the above text are from Reference 1 listed below which is published in Journal of Developmental Biology by MDPI.
References
- Navneet Dogra; Carmen Ledesma-Feliciano; Rwik Sen; Developmental Aspects of SARS-CoV-2, Potential Role of Exosomes and Their Impact on the Human Transcriptome. Journal of Developmental Biology 2021, 9, 54, 10.3390/jdb9040054.Bwire, G.M.; Njiro, B.J.; Mwakawanga, D.L.; Sabas, D.; Sunguya, B.F. Possible vertical transmission and antibodies against SARS-CoV-2 among infants born to mothers with COVID-19: A living systematic review. J. Med. Virol. 2021, 93, 1361–1369.
- Tallarek, A.C.; Urbschat, C.; Fonseca Brito, L.; Stanelle-Bertram, S.; Krasemann, S.; Frascaroli, G.; Thiele, K.; Wieczorek, A.; Felber, N.; Lütgehetmann, M.; et al. Inefficient Placental Virus Replication and Absence of Neonatal Cell-Specific Immunity Upon Sars-CoV-2 Infection During Pregnancy. Front. Immunol. 2021, 12, 698578.
- Tezer, H.; Demirdağ, T.B. Novel coronavirus disease (COVID-19) in children. Turk. J. Med Sci. 2020, 50, 592–603.
- Gaglani, S.; Gonzalez-Kozlova, E.; Lundon, D.J.; Tewari, A.K.; Dogra, N.; Kyprianou, N. Exosomes as A Next-Generation Diagnostic and Therapeutic Tool in Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 10131.
- Chen, T.Y.; Gonzalez-Kozlova, E.; Soleymani, T.; La Salvia, S.; Kyprianou, N.; Sahoo, S.; Tewari, A.; Cordon-Cardo, C.; Stolovitzky, G.U.S.T.A.V.O.; Dogra, N. Extracellular Vesicles Carry Distinct Proteo-Transcriptomic Signatures That are Different from Their Cancer Cell of Origin. bioRxiv 2021.
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514.
- Sabapatha, A.; Gercel-Taylor, C.; Taylor, D.D. Specific Isolation of Placenta-Derived Exosomes from the Circulation of Pregnant Women and Their Immunoregulatory Consequences. Am. J. Reprod. Immunol. 2006, 56, 345–355.
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977.
- Théry, C.; Duban, L.; Segura, E.; Véron, P.; Lantz, O.; Amigorena, S. Indirect activation of naïve CD4+ T cells by dendritic cell–derived exosomes. Nat. Immunol. 2002, 3, 1156–1162.
- Grommé, M.; Uytdehaag, F.G.C.M.; Janssen, H.; Calafat, J.; van Binnendijk, R.S.; Kenter, M.J.H.; Tulp, A.; Verwoerd, D.; Neefjes, J. Recycling MHC class I molecules and endosomal peptide loading. Proc. Natl. Acad. Sci. USA 1999, 96, 10326–10331.
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125.
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283.
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383.
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta (BBA) Biomembr. 1981, 645, 63–70.
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420.
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289.
- Greening, D.W.; Gopal, S.K.; Xu, R.; Simpson, R.J.; Chen, W. Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 2015, 40, 72–81.
- Greening, D.W.; Xu, R.; Gopal, S.K.; Rai, A.; Simpson, R.J. Proteomic Insights into Extracellular Vesicle Biology-Defining Exosomes and Shed Microvesicles. Expert Rev. Proteom. 2017, 14, 69–95.
- Hurley, J.H. ESCRT s are everywhere. EMBO J. 2015, 34, 2398–2407.
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17.
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208.
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30.
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of mRNAs and microRNAs is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659.
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron Microscopic Evidence for Externalization of the Transferrin Receptor in Vesicular Form in Sheep Reticulo-Cytes. J. Cell Biol. 1985, 101, 942–948.
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172.
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303.
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398.
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180.
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769.
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4.
- Farahani, M.; Rubbi, C.; Liu, L.; Slupsky, J.R.; Kalakonda, N. CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS ONE 2015, 10, e0141429.
- Driedonks, T.A.P.; van der Grein, S.G.; Ariyurek, Y.; Buermans, H.P.J.; Jekel, H.; Chow, F.W.N.; Wauben, M.H.M.; Buck, A.H.; Hoen, P.A.C.; Hoen, E.N.M.N. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cell. Mol. Life Sci. 2018, 75, 3857–3875.
- Gaglani, S.; Gonzalez-Kozlova, E.; Lundon, D.J.; Tewari, A.K.; Dogra, N.; Kyprianou, N. exRNA Signatures in Extracellular Vesicles and their Tumor-Lineage from Prostate Cancer. medRxiv 2020.
- Bai, K.; Li, X.; Zhong, J.; Ng, E.H.Y.; Yeung, W.S.; Lee, C.-L.; Chiu, P.C.N. Placenta-Derived Exosomes as a Modulator in Maternal Immune Tolerance During Pregnancy. Front. Immunol. 2021, 12.
- Neerukonda, S.N.; Tavlarides-Hontz, P.; McCarthy, F.; Pendarvis, K.; Parcells, M.S. Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek’s Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens. Genes 2019, 10, 116.
- Kharbanda, E.O.; Haapala, J.; DeSilva, M.; Vazquez-Benitez, G.; Vesco, K.K.; Naleway, A.L.; Lipkind, H.S. Spontaneous Abortion Following COVID-19 Vaccination During Pregnancy. JAMA 2021, 326, 1629.
- Nicolini, A.; Ferrari, P.; Biava, P. Exosomes and Cell Communication: From Tumour-Derived Exosomes and Their Role in Tumour Progression to the Use of Exosomal Cargo for Cancer Treatment. Cancers 2021, 13, 822.
- Marsh, M. The entry of enveloped viruses into cells by endocytosis. Biochem. J. 1984, 218, 1–10.
- Südhof, T.C.; Rothman, J.E. Membrane Fusion: Grappling with SNARE and SM Proteins. Science 2009, 323, 474–477.
- Mihalopoulos, M.; Dogra, N.; Mohamed, N.; Badani, K.; Kyprianou, N. COVID-19 and Kidney Disease: Molecular Determinants and Clinical Implications in Renal Cancer. Eur. Urol. Focus 2020, 6, 1086–1096.
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6.
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R. Beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e14.
- Qadir, F.; Aziz, M.A.; Sari, C.P.; Ma, H.; Dai, H.; Wang, X.; Raithatha, D.; Da Silva, L.G.L.; Hussain, M.; Poorkasreiy, S.P.; et al. Transcriptome reprogramming by cancer exosomes: Identification of novel molecular targets in matrix and immune modulation. Mol. Cancer 2018, 17, 1–16.
- Bland, C.L.; Byrne-Hoffman, C.N.; Fernandez, A.; Rellick, S.L.; Deng, W.; Klinke, D.J. Exosomes Derived from B16F0 Melanoma Cells Alter the Transcriptome of Cytotoxic T Cells that Impacts Mitochondrial Respiration. FEBS J. 2018, 285, 1033–1050.
- Giacomini, E.; Scotti, G.M.; Vanni, V.S.; Lazarevic, D.; Makieva, S.; Privitera, L.; Signorelli, S.; Cantone, L.; Bollati, V.; Murdica, V.; et al. Global transcriptomic changes occur in uterine fluid-derived extracellular vesicles during the endometrial window for embryo implantation. Hum. Reprod. 2021, 36, 2249–2274.
- Esfandyari, S.; Elkafas, H.; Chugh, R.M.; Park, H.-S.; Navarro, A.; Al-Hendy, A. Exosomes as Biomarkers for Female Reproductive Diseases Diagnosis and Therapy. Int. J. Mol. Sci. 2021, 22, 2165.
More
