The human kallikrein-related peptidase 4 (KLK4) and the transcribed pseudogene KLKP1 are reported to be highly expressed in the prostate. When trying to clone transcripts of KLKP1, authors partly failed. Instead, they identified an androgen-regulated transcript, KLK4T2, which appeared to be a splice variant of KLK4 that also contained exons of KLKP1. Expression analysis of KLK4, KLK4T2, and KLKP1 transcripts in prostate cancer cell lines showed high levels of KLKP1 transcripts in the nucleus and in unfractionated cell extract, whereas it was almost completely absent in the cytoplasmatic fraction. This was in contrast to KLK4 and KLK4T2, which displayed high to moderate levels in the cytoplasm. In patient cohorts we found significantly higher expression of both KLK4T2 and KLK4 in benign prostatic hyperplasia compared to both primary prostate cancer and bone metastasis. Analysis of tissue panels demonstrated the highest expression of KLK4T2 in the prostate, but in contrast to the classical KLK4, relatively high levels were also found in placenta. So far, the function of KLK4T2 is still to be explored, but the structure of the translation product indicated that it generates a 17.4 kDa intracellular protein with possible regulatory function.
1. Introduction
Prostate cancer (PC) is a major global medical problem, with an annual incidence of close to 1.5 million cases in 2020 (Global Health Observatory) and in Sweden it is the most frequently diagnosed tumor disease among men with about 11,000 new cases per year (Swedish Cancer Registry). PC is a multifaceted disease, and it is not possible today to predict whether a tumor will develop into an aggressive form or not. The mechanisms behind PC metastasis and castration-resistant tumor growth are largely unknown and identification of molecular pathways involved has the potential to improve both PC prognosis and treatment.
The kallikrein-related peptidases, KLKs, are a family of serine proteases, which in humans are encoded by 15 genes situated at a locus on the long arm of chromosome 19
[1][2][3]. Aberrant expression of KLKs has been implicated to be involved in different stages of cancer development. Cleaving molecules and remodeling the extracellular matrix, activation of prohormones, and/or signaling molecules, leads to increased cell proliferation and angiogenesis, which contributes to tumor growth, invasion, and metastasis formation
[4][5][6][7][8]. Several studies have also been undertaken to analyze their potential as biomarkers of various cancers, as reviewed in
[9][10][11].
It is also known as prostate-specific antigen (PSA) and is currently widely used as a marker for PC detection and disease development. KLK2, which is located next to and is closely related to KLK3, has also been shown to be an excellent PC marker
[12]. There are also reports suggesting a role for both KLK2 and KLK3 in the etiology of prostate cancer
[13][14]. Another neighbor of KLK2 is KLK4, which was first cloned as enamel matrix serine proteinase 1 (EMSP1) and shown to be important for tooth enamel formation
[15]. In parallel, the transcript was cloned and analyzed by other groups under the name prostase, KLK-Like 1, PRSS17, androgen-related message 1 (ARM1), and KLK4
[16][17][18][19]. Study of the gene showed that it was organized with six exons, the first of which containing 5′ nontranslated nucleotides only
[18]. There was also a study demonstrating that KLK4 differed from other KLKs, by primarily being synthesized without signal peptide and thus becoming an intracellular protein in the prostate
[20].
Between KLK2 and KLK4 on human chromosome 19, there is a gene with the HGNC-approved designation KLKP1. Originally it was identified as a transcribed pseudogene containing a nucleotide sequence homologous with the second translated exon of KLK1
[21]. It has since then been described as a gene generating the noncoding transcript KLK31P, but also of coding transcripts yielding the product KRIP1
[22][23]. These studies also showed that the gene is downregulated during prostate cancer progression.
2. Identification of a Transcript from the KLKP1 Locus as a Splice Variant of KLK4
Initially, we intended to clone the KLK31P transcripts (GenBank accession number DQ211698 and AY533562) and the transcripts encoding KRIP1 (GenBank accession number DQ086829), using RNA isolated from BPH tissue and LNCaP cells as template for RT-PCR. Despite repeated efforts, we failed to generate the KLK31P transcripts. We were more successful with the KRIP1 transcript, but in addition to the expected transcript, the RT-PCR also generated a smaller product in which nucleotides 358–787 of DQ086829 were missing. As there were GT and CAG sequences at the ends of the missing piece, it most likely was removed by a splicing event, which also, almost completely, removed the nucleotides coding for KRIP1.
The difficulty in obtaining the KLK31P transcripts in combination with our discovery of a new transcript from the KLKP1, prompted us to reinvestigate the products of the gene by RACE analysis. We used the primer KLKP9R, originally synthesized for cloning of KLK31P and KRIP1, in 5′ RACE experiments (
Table 1). To rule out heterogeneity in the 3′ end, we also made 3′ RACE with an oligonucleotide, priming to a common sequence of KLK31P and KRIP1, which is also homologous with exon 2 of KLK1. The results of the RACE experiments are shown in
Figure 1. The 3′ RACE yielded a single 0.8 kb product, with a nucleotide sequence identical to previously reported transcripts, and ending in a poly(A) tail showing it was derived from an mRNA. The 5′ RACE yielded a single product of 1.2 kb, suggesting a single predominating product from the gene. The nucleotide sequence of the 3′ part of the transcript was identical to the one we generated by 3′ RACE. Upstream of the sequence homologous with the second exon of KLK1, we expected to find the sequence present in both KLK31P and KRIP1. To our surprise, this was not the case. Instead there was a sequence that we could identify as coming from exon 4 and 3 of KLK4 and with a 5′ terminus that was very similar to the one reported for the transcript of the intracellular form of KLK4
[20]. Thus, the major product of the KLKP1 locus seemed to be a splice variant of KLK4. We named this transcript KLK4T2.
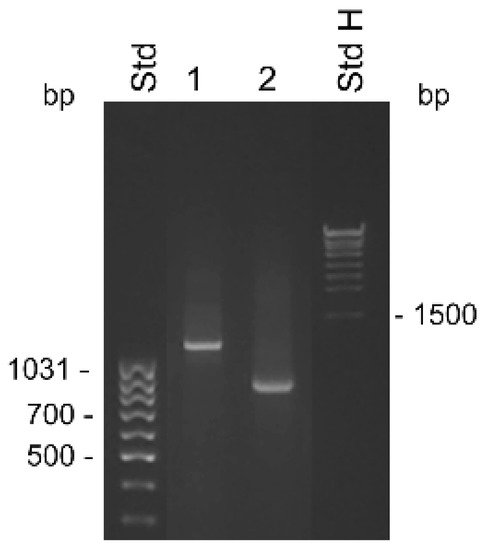
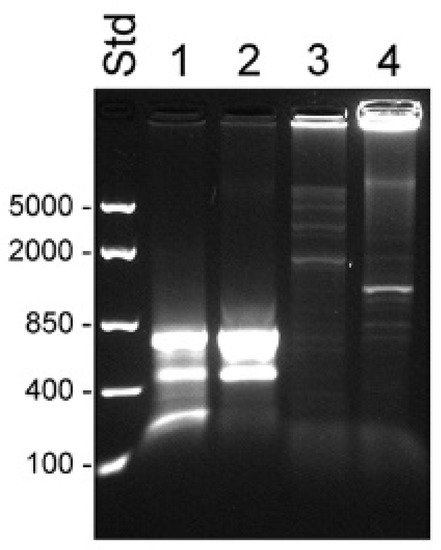
Figure 1. Detection of KLK4T2 by RACE. RACE products generated from 2 ng unfractionated LNCaP cell RNA were analyzed by electrophoresis on 1% agarose gel. Lane 1 shows 5′ RACE with the specific primer KLKP9R and lane 2 shows 3′ RACE with the specific primer KLKP8F. For information about the primer sequences, see Table 1. MassRuler Low Range (Std L) and High Range (Std H) were used as molecular size markers. Relevant sizes in bp are indicated in the figure.
Table 1. Primers used for PCR, cloning, and DNA sequencing. Bases written in lower case are non-sequence-specific nucleotides included for the cloning procedure.
| Primer |
Sequence 5′–3′ |
Purpose |
| KLKP9R |
tggcagcggccgcAACATCACAGCTTCTGTTTATTAAGCA |
5′ RACE, cloning |
| KLKP8F |
tcgcagaattcATGCTGTGATTGCCATCCAGTCCCAGA |
3′ RACE, cloning |
| KLK4r18 |
gatcggtcgactgACTGGCCTGGACGGTTTTCTCTAT |
5′ RACE, cloning |
| KLKPf10 |
agctgaattcTAAACGGCGAGGACTGCAGCCCGCACT |
cloning |
| KLK4Ne2f |
GGCGGCACTGGTCATGGAAAACGAA |
3′ RACE |
| KLK4Ne3f |
CGGAGCATCAGCATTGCTTCGCAGT |
3′ RACE |
| K4T2e23f |
CTGCTGGCGAACGATGCTGT |
qPCR |
| K4T2e3r |
GACGTCCTTGCACTGGAAGT |
qPCR |
| KL4e34f |
CTGCTGGCGAACGGCAGAAT |
qPCR |
| KL4e4r |
GTGGTACAGCGGGTCATAGA |
qPCR |
| KPT2e1f |
ACTCCCTCTGCAGATGCTGT |
qPCR |
| KLKPF1 |
CCCAACCCTGGCAGGGTTGTACCATT |
DNA sequencing, cloning |
| KLKPR4 |
ACTTCGGAGAACTATGGTGCTGGCTA |
DNA sequencing |
| KLK4Ne4r |
AGCTTACTGCAGACCTCCTCAGACA |
qPCR |
| HPRTf1 |
AGGGGACATAAAAGTAATTGGTGGAGAT |
qPCR |
| HPRTr2 |
TGACCAAGGAAAGCAAAGTCTGCATTGT |
qPCR |
| rpl13a-f |
GTACGCTGTGAAGGCATCAA |
qPCR |
| rpl13a-r |
GTTGGTGTTCATCCGCTTG |
qPCR |
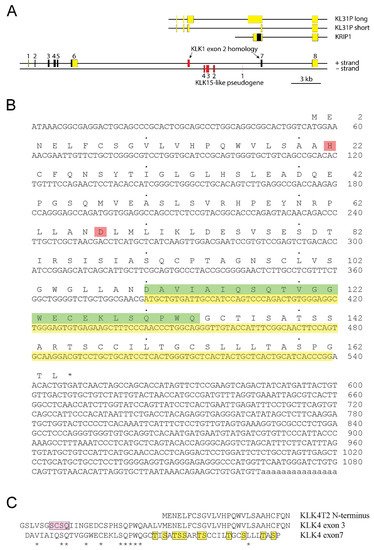
Our result also suggested that KLK4 was a fairly large gene, consisting of at least 8 exons (
Figure 2A). The structure of KLK4T2 cDNA with its translation is shown in
Figure 2B. The transcript did not encode a signal peptide and therefore, the product presumably was a small intracellular protein with a predicted molecular mass of 17.4 kDa. The His and Asp of the catalytic triad were preserved in the KLK4T2 product, which most likely was lacking proteolytic activity, as the catalytic Ser, located in exon 5 of the gene encoding the KLK4 protease, was missing.
Figure 2. The extended KLK4 locus. (A) Schematic drawing of the KLK4 gene with location of previously reported KLKP1 genes illustrated above. Exons are indicated by boxes with translated sequences shown in black and nontranslated sequences in yellow. Exons of pseudogenes are colored in red. The new KLK4T2 transcript is formed by exon 4, 7, 8, and part of exon 3. (B) The cDNA sequence of KLK4T2 is shown with translation in one letter code written above. Stop codon is marked with an asterisk. The His and Asp part of the catalytic triad in KLK4 are marked in pink. Nucleotide sequence homologous to KLK1 exon 2 is highlighted in yellow, and the corresponding amino acid sequence until the frame shift, in green. (C) Conservation of amino acid residues encoded by exons 3 and 7 in KLK4. Amino acid alignments showing conserved residues encoded by KLK4 exon 3 and 7 indicated by stars. The pro-piece of proKLK4 is highlighted in pink and the Ser and Thr residues in the C-terminal part of the KLK4T2 product are highlighted in yellow. The N-terminal part of the KLK4T2 product is shown above the aligned sequences for comparison.
An interesting observation was made when aligning the primary structures encoded by exon 3 and exon 7 of the extended KLK4. There were conserved residues that reached approximately halfway into the exons, as seen in
Figure 2C. This region of the KLK4 protease holds the residues of the pro-piece and the amino terminus of the activated protein.
3. Analysis of Transcripts from the KLK4/KLKP1 Locus by RACE
The 5′ RACE experiments were run on samples made with total RNA from BPH tissue and LNCaP cells and fractionated RNA isolated from the cytosol and nucleus of LNCaP cells. Priming with oligonucleotide KLKP9R, which hybridized to the 3′ end of KLK4T2 and also of KLK31P and KRIP1, yielded a single predominating product of 1.2 kb in all samples (
Figure 3). Sequencing of the product showed that it was identical to KLK4T2.
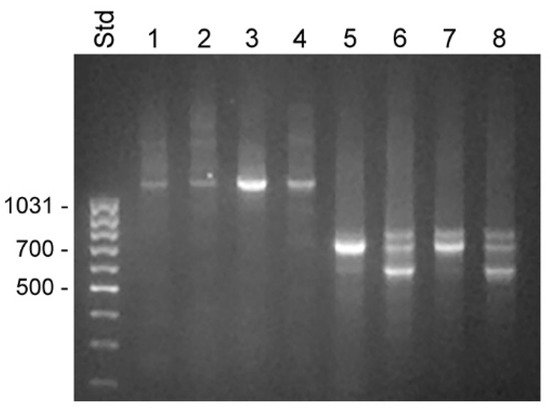
Figure 3. Transcripts detected by 5′ RACE in unfractionated BPH and compartments of LNCaP cells. RACE products were analyzed by electrophoresis in 1% agarose. Lanes 1–4 show 5′ RACE experiments using the KLK4T2-specific primer KLKP9R and lanes 5–8 using the primer KLK4r18, specific for classical and truncated KLK4. Products generated with unfractionated RNA from BPH are shown in lanes 1 and 5 and from LNCaP in lanes 2 and 6. Products generated with RNA from the cytosol of LNCaP are shown in lanes 3 and 7, and from the nucleus in lanes 4 and 8. MassRuler Low Range (Std) was used as size marker and relevant sizes in bp are written to the left.
The primer KLK4r18, which hybridized to a sequence immediately upstream to the stop codon in KLK4, resulted in a more complex pattern, with a 0.7 kb product that was present in all samples (
Figure 3). In the BPH sample, it appeared as a single predominating product, but it was also the major product in the sample from LNCaP cytosol. DNA sequencing identified it as a KLK4 transcript, identical to that described by
[20], with a 5′ end very similar to that of KLK4T2, but around 30 bp shorter. The dominating component in LNCaP samples made from nucleus or total RNA was a 0.6 kb product. The sequencing of the cDNA showed that it had the expected 3′ end in exon 6, but then the sequence continued into intron 5, where it after 319 bp ended in a polyA tract. A third component was also evident in all LNCaP samples. The cDNA sequence of this 0.8 kb product showed it to be identical to the 0.7 kb component, but also carrying the fourth intron of KLK4, which had not been spliced out. Both the 0.6 and 0.8 kb products could indicate that the splicing machinery in the LNCaP cells is less efficient.
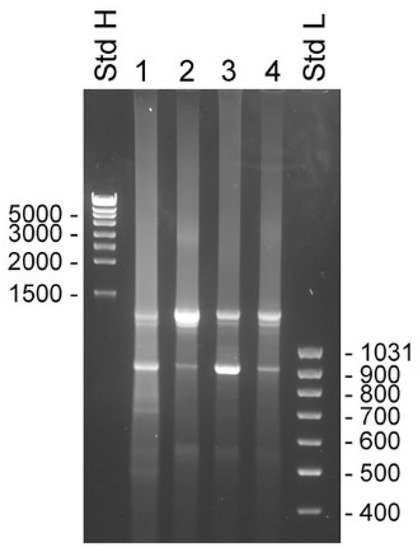
The 3′ RACE experiments were made with samples taken from the same sources as used for the 5′ RACE experiments. The primers used were KLK4Ne2f and KLK4Ne3f, which hybridized with sequences in KLK4 exon 3 and 4, respectively. Both primers gave very similar results, with the exception that priming in exon 3 yielded slightly larger products that reflected the 262 bp difference in location of priming sites. There were two major components present in all samples. The estimated molecular sizes of products generated by KLK4Ne3f were 1.3 and 0.9 kb (
Figure 4). The cDNA sequence showed that the smaller product emanated from KLK4T2, which was predominant in samples made with total RNA from BPH and LNCaP cytosol. The larger product was predominant in samples made with total RNA and the nucleus fraction of LNCaP cells. The sequence of the product generated from total RNA of LNCaP yielded a sequence extending from exon 4 to exon 5 and then continued into intron 5, where it ended in a poly(A) sequence located 935 bp into the intron. Sequencing of the upper band from BPH prostate and cytosol of LNCaP yielded unreadable sequences, presumably due to heterogeneity of the 3′ RACE products.
Figure 4. Transcripts detected by 3′ RACE in unfractionated BPH and compartments of LNCaP cells. RACE products were analyzed by electrophoresis in 1% agarose gels. Experiments were done with the primer KLK4Ne3f, which is specific for KLK4, exon 4, which will generate product from both classical and truncated KLK4, and KLK4T2. The 3′ RACE-ready cDNA was synthesized with unfractionated RNA from human BPH tissue (lane 1) and LNCaP cells (lane 2), and RNA from LNCaP cytosol (lane 3) and nucleus (lane 4). MassRuler high (Std H) and MassRuler low (Std L) were used as size markers. Relevant sizes in bp are indicated in the figure.
The 5′ RACE cDNA was also synthesized from pooled material consisting of total RNA prepared from bone metastasis of patients suffering from castration-resistant prostate cancer (CRPC) or hormone naïve prostate cancer (HNPC) (
Figure 5). Using the KLK4-specific primer KLK4r18, three products were formed from the CRPC and two from the HNPC material (lanes 1 and 2). The largest, 0.7 kb product, corresponded well with the sequence published by Korkmaz et al.
[20], starting 20–40 bases into exon 3, followed by exons 4–6. Some of the fragments contained an extra 12 bp insertion between exon 3 and 4. The 0.6 kb product seemed to be a splice variant resembling the larger product except it totally lacked exon 5. The third and smallest band from the CRPC turned out to be a mix of primer dimer and nonidentified short sequences. Several diffuse, high molecular weight bands were formed when using the KLK4T2-specific primer KLKP9R in 5′ RACE PCR using CRPC as template, but no clones could be isolated and sequenced from this material (
Figure 5, lane 3). When using HNPC as template, a weak 1.2 kb fragment was formed that corresponded to KLK4T2. In the CRPC sample, but especially when using HNPC as template, we also obtained amplification of fragments so large that they could hardly enter the gel (
Figure 5, lanes 3 and 4).
Figure 5. Transcripts detected by 5′ RACE PCR in RNA from prostate cancer bone metastasis. RACE products were separated on a 1.2% agarose gel. The 5′ RACE cDNA was synthesized using pooled total RNA prepared from bone metastasis of patients suffering from castration-resistant (lanes 1 and 3, n = 5) or hormone-naïve (lanes 2 and 4, n = 2) prostate cancer. PCR was run using KLK4-specific oligo KLK4r18 (lanes 1 and 2) or KLK4T2-specific oligo KLKP9R (lanes 3 and 4) in combination with the UPM primer from the RACE kit. RNA was prepared using AllPrep DNA/RNA/Protein kit (Qiagen). FastRuler Middle Range (Std) was used as molecular weight standard, and relevant sizes in bp are indicated in the figure to the left.
4. Analysis of Transcripts from the KLK4/KLKP1 Locus by RT-qPCR
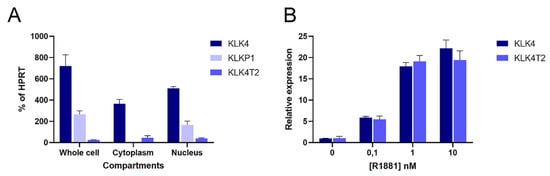
Because of the discovery in the RACE experiments, we decided to quantify transcripts from different compartments of the combined KLK4 and KLKP1 loci. We used primers that would specifically recognize KLK4, KLK4T2, and KLKP1 (KRIP1 and the longer form of KLK31P) in RT-qPCR experiments. This showed that the transcripts encoding KLK4 and KLK4T2 were found in the cytosol fraction (
Figure 6A). In contrast, the KLKP1 transcripts were enriched in the nuclear fraction, while it was more or less eliminated in the cytoplasmatic fraction.
Figure 6. Cellular location and androgen stimulation of transcripts. Transcripts were measured by RT-qPCR in extracts of LNCaP cells. PCR was run using the primer pairs KLK4Ne3f-KLK4Ne4r amplifying KLK4, and K4T2e23f-K4T2e3r specific for KLK4T2. KPT2e1f-K4T2e3r used for KLKP1 amplifies both KRIP1 and KLK31P long transcripts. The results are given as concentration relative to the transcript concentration of the housekeeping gene HPRT, which was amplified using HPRTf1-HPRTr2. The error bar represents one standard deviation. (A) The relative amounts of KLK4, KLK4T2, and KLKP1 transcripts in total (whole cell compartment) or fractionated RNA (Cytoplasm or Nucleus) are shown. (B) To explore the androgen sensitivity, RT-qPCR was run on RNA prepared from LNCaP cells induced with 0.1, 1, and 10 nM R1881, as indicated in the figure. The amount of KLK4 and KLK4T2 transcript are expressed relative to noninduced levels for each species.
Hormone effect on the KLK4T2 expression was studied in androgen-stimulated LNCaP cells. The androgen stimulation resulted in an increase of KLK4T2 transcripts, which was of a similar magnitude as the hormone-induced increase of KLK4 (
Figure 6B).
5. Expression Pattern of KLK4-Related Transcripts in the Prostate and during Prostate Cancer Disease Progression
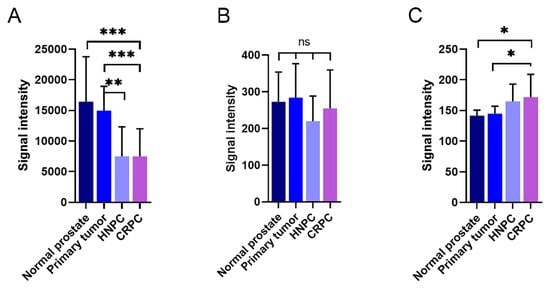
In Illumina expression array analysis, two different probes were hybridizing with KLK4-related transcripts, denoted KLK4 and KLKP1. As can be seen in
Figure 7A, the expression of KLK4 was very high in both normal and primary prostate cancer tissue. When the disease progressed, the transcript levels of KLK4 were significantly reduced, as can be seen from the levels in both hormone-naïve and castration-resistant bone metastases. This was in contrast to the findings for KLKP1, where no significant differences could be observed between the different disease stages (
Figure 7B). Contrary to KLK4, the Ki67 expression, representing tumor cell proliferation rate, was as expected, positively correlated to the disease stage (
Figure 7C).
Figure 7. Expression of KLK4-related transcripts in patient materials measured using an Illumina whole genome expression array. The amount of KLK4-related transcripts designated as KLK4 (A) and KLKP1 (B) were analysed in normal prostate, primary prostate cancer tumours, and bone metastasis from patients suffering from hormone-naïve (HNPC) or castration-resistant prostate cancer (CRPC), respectively. The corresponding results for the proliferation marker Ki67 are shown in (C). The error bars represent one standard deviation. *** p < 0.001, ** p < 0.01, * p < 0005, ns = not significant.
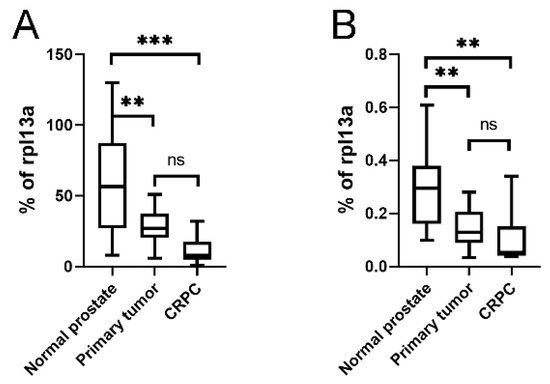
As the KLKP1 probe used in the Illumina array showed 100% sequence identity with both KLK4T2 as well as several other published KLKP1 variant sequences, we went on and ran RT-qPCR on patient samples using primer pairs specific for KLK4 and KLK4T2. In
Figure 8, the expression pattern in normal prostate tissue (
n = 10), primary tumor tissue (
n = 13), and bone metastasis from CRPC (
n = 9) are displayed. As can be seen, the expression of KLK4 was significantly decreased as the disease progressed, although the most significant difference is seen as the disease progressed into bone metastasis (
Figure 8A). The expression of KLK4T2 (
Figure 8B) was also decreased in primary tumor and bone metastasis compared to normal tissue.
Figure 8. Expression of KLK4 and KLK4T2 transcript in patient material. Transcript levels were measured by RT-qPCR on unfractionated RNA prepared from normal human prostate tissue (n = 10), primary PC tumor (n = 13) or bone metastasis from CRPC patients (n = 9). In the qPCR, the primer pairs KLK4Ne3f-KLK4Ne4r amplifying KLK4 (A) and K4T2e23f-K4T2e3r specific for KLK4T2 (B) were used. The results are given as concentration relative to the transcript concentration of the housekeeping gene rpl13a, which was amplified using primer pairs rpl13a-f–rpl13a-r. Samples were run as triplicates, and data are plotted as boxplots with the median value as a line across the box and the first quartile (Q1) value at the bottom and the third (Q3) at the top. The whiskers show the highest and lowest values in each group. *** p < 0.001, ** p < 0.01, ns = not significant.