Extracellular vesicles (EVs) are 50–1000 nm vesicles secreted by virtually any cell type in the body. They are expected to transfer information from one cell or tissue to another in a short- or long-distance way. RNA naturally present in EVs might be limited in a physiological context.
- extracellular vesicles
- exosome
- RNA
- miRNA
1. Introduction
2. RNA-Based Mechanism of Action for Native EVs
Many teams reported an (mi)RNA-based mechanism of action for EVs. Most proofs of concept are however in vitro, and in vivo data are relatively scarce or not designed to claim an RNA based mechanism of action delivered via EVs at long distances and/or limited to short-distance transfer (Table 1):-
In glioblastoma (GBM), Abels et al. describe short distance communication through EVs from tumor cells to microglia to induce microglia reprogramming. The presence of EVs was detected in 0.3% of microglial cells, and the presence of the miRNA of interest transferred by EVs was detected in these sorted 0.3% of cells. However, no clear target protein silencing was found (only 4 out of 59 validated targets). It may be possible that the miRNA detected would partly be coming from the retention of EVs (and its associated miRNA) in endosome of microglial cells, without intracytosolic delivery [19][2].
-
Lucero et al. demonstrate the short distance effect of glioblastoma-derived EVs to induce angiogenesis via miRNAs in vitro, and claim it to be also valid in humans only based on a correlation with a human glioblastoma transcriptomic “fingerprint” [20][3]. However, no clear demonstration of causality is proposed.
-
Shen et al. demonstrate the effect of EVs derived from tumors to induce stemness via miRNA in surrounding cells in vitro (at supra physiologic doses) and claim it to be also valid in vivo in tumor-bearing mice. However, they used Rab7 KO tumors as a control to inhibit EV production, a KO that also has a lot of other side effects [21][4]. It is therefore difficult to know whether this effect is mediated by EVs and by the miRNA inside them.
-
Ying et al. demonstrate a role for miR-155 transferred by EVs in vitro in glucose tolerance and use an elegant system of bone marrow transplantation to investigate the role of hematopoietic derived miR-155 in a KO mouse. They later claim that the partial rescue of physiologic glucose tolerance is mediated by EVs in vivo although it may also be mediated by other intercellular transfer mechanisms like tunneling nanotubes (TNT), especially to transfer at short distance miRNA from a very macrophage-rich organ like liver to surrounding hepatocytes [22][5]. The same miRNA-155 has indeed been shown to be able to be transferred through TNT [23][11].
-
Thomou et al. help us to raise other non-trivial questions on vesicular versus non vesicular mediated RNA transfer. He proposed that EVs from adipose tissue would be able to transfer miRNA to liver cells and induce RNA silencing in vivo. The protein expression is reduced by up to ∼95% after injection of serum-derived EVs (from donor mice with brown adipose tissue expressing the miRNA of interest) to miR-KO mice [24][7]. Strictly speaking, the demonstration proves that a serum factor purified with common EV purification protocols from the donor mice leads to specific miRNA-mediated silencing in mice. It raises the question of whether this effect may be at least partly mediated by an extra-vesicular miRNA in serum co-purified with EVs.
-
Other teams claimed the demonstration of an efficient transfer of CRE-mRNA via EVs [27,28,29][8][9][10] in vivo. This highly sensitive “on/off” system induces or stops the expression of a particular fluorescent protein upon delivery of the CRE-recombinase protein or its RNA. Although it is very different from a physiologic system, it may still be of interest as a proof of concept. However, this assay has shown limited transduction efficacy even with a high dose of EVs (e.g., in Ilahibaks et al. [30][12] achieved ∼15% transduction efficacy by ∼8300 EV/cell in vitro, i.e., intra-cytosolic transfer of at least one CRE protein or RNA). More importantly, it may be biased by the transfer of a single CRE recombinase protein (instead of CRE-mRNA) from the donor EVs, although it was not detected in these articles. On the contrary other teams clearly reported the presence of CRE protein in EVs produced from CRE-producing cells [31][13].
| Author | Model/Context | Long/ Short Distance |
Demonstration | Limits |
|---|---|---|---|---|
| Abels et al. [2] |
Glioblastoma (GBM) | Short | Transfer of GBM EVs in 0.3% of microglial cells, presence of a GBM-miRNA in these cells | Partial (4/59) miRNA target induced silencing |
| Injection of GBM EVs induce partial siRNA target knockdown | Highly supra-physiologic EV dose | |||
| Lucero et al. [3] |
Glioblastoma (GBM) | Short | GBM EVs induce angiogenesis in vitro and a transcriptomic fingerprint is described | Supra physiologic dose (105 EVs/cell) |
| The transcriptomic fingerprint is found is also found in patients | There is only a correlation between EV treated GBM cells and patient GBM, no demonstration of causality is proposed | |||
| Shen et al. [4] |
Tumor derived EVs | Short | Tumor derived EVs induce stemness in vitro | Supra physiologic dose (25:1 producing to receptor ratio) |
| Limiting EV transfer in vivo diminish the effect on surrounding cells | KO of EV production in vivo is performed using a Rab-7 KO tumor model, this KO has a lot of other effects that may explain the difference observed | |||
| Ying et al. [5] |
Glucose tolerance | Potentially both | miRNA is transferred from hematopoietic derived cells to liver cells in vivo | The transfer may be mediated by either EVs or Tunelling nanotubes (TNT) (and other?) mechanism, this miRNA being known to be transferred via TNT |
| Chen et al. [6] |
Bone regeneration | Potentially both | MiR-375 is able to induce bone regeneration in vitro | No significant difference is observed compared to EVs not expressing miR-375 in vivo |
| Thomou et al. [7] |
Transfer of miRNA from adipose tissue to liver | Potentially both | A serum-derived EV preparation transfers active miRNA to liver cells in vivo | The serum derived EV preparation purification protocol has a high chance to be comtaminated by extravesicular miRNA (up to 97.5% of miRNA purified) |
| Various teams [8][9][10] |
CRE-mRNA transfer in vivo | Potentially both | CRE recombination is induced at long distance in the presence of EVs derived from cells expression CRE mRNA and protein | The CRE-Lox induced recombination may be mediated either by mRNA transfer via EVs but also or by transfer of mRNA or CRE protein by TNT, cell fusion, or extravesicular transfer |
| A particular phenotype is described in CRE-recombined cells compared to non recombined cells | The causality in not demonstrated as a cell with a particular phenotype may be more prone to be transfected by CRE, in particular a more mobile and phagocytic cell. The sole endocytosis of nano-objects like EVs is also impacting the cell phenotype, even in the absence of cargo. |
3. Physiological Effect of RNA Cargo in EVs: A Natural RNA Vector?
3.1. Stochiometric Evaluation of RNA Loading in EVs
Sverdlov claimed that it is very unlikely that naturally circulating EVs transfer a significant part of information through RNA in vivo at long distances in physiological states [14][18]. He argued that the best candidates for information transfer would be self-amplifying (e.g., mRNA) and/or have a regulatory function (e.g., a transcription factor, a miRNA). At the time, he made the hypothesis that RNA inside EVs was not subject to strong selection. Baglio et al. [38][19] and other groups found that most RNA in various types of EVs (from tumor, MSCs, immune cells and serum, isolated by various methods (ultra)-centrifugation or affinity column) were small <400 nucleotides (nt) long RNA [39,40,41,42][20][21][22][23]. Among them, most are tRNAs (that can hardly be expected to have an effect) and miRNA only constituted ∼0.9% [43][24] of RNA reads. Although the miRNA are relatively enriched (∼10 fold compared to cell RNA4), enrichment may largely be due to the nonspecific size selection biased to the smaller sizes such as tRNAs. As an example, 16 S RNA (1,6 kB), a typical medium-size RNA has a hydrodynamic diameter of ∼30 nm [44][25], whereas miRNA (20–83 nts) have a cylinder shape with a 2 nm diameter and a 7–20 nm length. mRNA encapsulation inside EVs also depends on their local concentration around EV formation sites, as well as mRNA interaction with membrane lipids and proteins [45][26]. Before being functional, miRNA are getting through the pri- and pre-miRNA state. To be potentially active if they get to the target cell cytosol, miRNA needs either (i) to be not yet associated with Ago2 to form the RISC complex but still able to bind to it (i.e., being pri- or pre-miRNA) and therefore they would be able to bind it later on in the recipient cell cytosol or (ii) to already be associated with the RISC complex as a miRNA, a state in which they can exert their silencing activity directly. Importantly, association of miRNA to the RISC complex allows them to be much more stable than if left alone where it can be rapidly degraded by nuclease, in particular in the context of EV travel through endosomes (containing nucleases) in the target cell.
Sverdlov proposed a rough approximation of the maximal amount of RNA per EV if they are densely packed in EV of 100 nm diameter: ∼1600 RNA/EV for 1000-nt RNA and ∼6700 RNAs/EVs for 200 nts RNA. However, when measured by total RNA quantification [48][27], the number of RNA per EV was less than one in serum-derived EVs. Another team reported the presence of ∼7 µg of RNA per 1010 EVs dosed by bulk representing ∼6500 RNA molecules per EV [43][24], but the presence, as discussed by the authors, of contaminating surrounding extra-vesicular RNA may artificially enhance this number. As an example, once extra-vesicular RNA is removed from serum-derived EV preparations (using differential centrifugation and size-exclusion chromatography) only ∼2.5% of total miRNA remains in the serum-derived EV fraction [25,26,49,50,51,52][28][29][30][31][32][33]. Most of the time, purification strategies used are not allowing complete extra-vesicular RNA removal (in particular in serum where it represents a large fraction of RNA), therefore attribution of a particular effect to intra-vesicular EVs may be difficult. Quantitative results on the amount of miRNA per EVs estimates that most represented miRNA can hardly be found in 1 out of 100 exosomes (the range varies for each miRNA from one copy per 9 exosomes to one copy per 47,162 exosomes, mean of 1 copy per 121 exosomes using digital PCR, a reliable and sensitive quantitative method) [25][28]. Knowing that they detected 131 miRNA in total, the estimated miRNA per EV should be considered to be ∼1 per EV.
3.2. Navigating the Bloodstream and Getting to the Target?
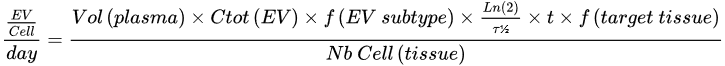
| Parameter | Proposed Value | Reference | |
|---|---|---|---|
| Ctot (EV) | 1012 EV/L | [37][38] | |
| f (EV subtype) | All EVs | 100% | [39] |
| Erythrocyte | 4% | ||
| Platelet | 51% | ||
| B cell | 25.7% | ||
| CD4 cell | 11% | ||
| All non hematopoietic tissue EVs | 0.2% | ||
| Adipose tissue | 0.16% | ||
| Other non hematopoietic tissue | 0.04% | ||
| Half life (τ½) | |||
| 7 min (mice) | [40] | ||
| f (target tissue) | All tissues | 100% | [36] |
| Liver | 60% | ||
| Spleen | 15% | ||
| Lung | 10% | ||
| Brain | 0.5% | ||
| Nb Cell (tissue) | All tissues | 3.72 × 1013 | [41] |
| Liver | 2.41 × 1011 | ||
| Spleen | 2 × 1011 | ||
| Brain | 3 × 1012 | ||
3.3. A Very Interesting Intra-Cytosolic RNA Delivery (Endosomal Escape)
3.4. Is the Physiologic RNA in EVs Dose Sufficient to Achieve an Effect?
The RNA-based mechanism of action (MOA) for effects mediated by non-modified native EVs in therapeutic conditions has previously been challenged by comparing it to data obtained from siRNA experiments. In most preclinical studies, EV doses usually range from ∼1 to 200 µg per mouse [81][51], corresponding to about 1010 to 1012 EV/mouse depending on EV preparation and dosage methods. If we consider ∼1 miRNA per EV, this dose represents ∼1010 to 1012 miRNA per dose, corresponding to about ∼0.2–20 ng of miRNA/mouse or ∼0.016–1.6 pmol/mouse. siRNA doses reported to be efficient in vivo in systemic injections are rather in the microgram range (27 to 750 µg/mouse [82,83][52][53]). One explanation is that the observed therapeutic effect of native EVs is not mediated by their naturally loaded (mi)RNAs. Indeed, this ∼103–104 fold difference was though too big to be explained by a very high difference in delivery efficacy [43,80][24][54]. However, this may be now discussed in view of recent results comparing engineered EVs to synthetic RNA nanovectors.
Indeed, recently reported delivery efficacy of EVs obtained in vivo show a ∼10–300 fold improvement in favor of EVs [84][55] compared to lipid nanoparticles (although the authors discuss the estimation of miRNA concentration with their method may favor EV reported efficacy by ∼10 fold [85,86][56][57]). The authors used the natural ability of pre-miR-451 to be enriched preferentially in EVs and used it as a backbone to couple with an siRNA of interest in order to target it inside EVs [84][55]. They then used these engineered EVs to target the liver, intestine or kidney glomeruli and achieve various target knockdown. Interestingly, this ∼10 to-300 fold improvement in terms of RNA cytosolic delivery in favor of EV in vivo is fully consistent with independent data on delivery efficacies reported for synthetic vectors: EVs reach a ∼20% endosomal escape rate [71][46] compared to 0.1 to 2% for synthetic vectors [87][58], which leads to a ∼50 fold increased cytosolic delivery. Even higher differences (up to 104) were reported in the delivery efficacy in favor of EVs in vitro [32][59]. Importantly, such a fold change also takes into account the very different endocytosis rate that favors EVs compared and synthetic vectors in vitro but not in vivo [88][60].
Altogether, these quantitative estimates show (Figure 1) that distant communication by EVs via RNAs probably has limited efficacy in physiological conditions, although it may be a bit different in pathological conditions and in the therapeutic use of EVs that are engineered to load large amounts of specific RNA.

4. Considerations on RNA Based Information Transfer in Therapeutic Settings
4.1. Considerations on the Therapeutic Effect of RNA from Unmodified EVs
4.2. Considerations on the Effect of RNA from Engineered EVs
Although unmodified EVs may difficultly have an effect via their RNA cargo, EVs may be very interesting vectors for synthetic delivery of exogenously loaded RNA (mi/si/mRNA). Exogenously EV-loaded synthetic RNA delivery for therapeutic purposes have been reported to treat various pathological conditions [101,102][62][63]. The problem of efficient EV loading is still highly debated, in particular in the case of electroporation [103][64] and was recently reviewed elsewhere by our team and others [11,104][65][66]. Briefly, loading of nucleic acid in EVs in EVs have been reported with various techniques ranging from electroporation [103][64], EV destruction-reformation techniques (slicing, extrusion or sonication [105][67]), permeabilization via saponin, the use of commercial transfection reagents like lipofectamine [106][68], heat shock [107][69], pH gradient [108][70], etc. Importantly, most of these methods suffer from a low loading efficiency, potential substantial denaturation of EVs and whether these methods lead to intra-vesicular loading or extra-vesicular aggregation on EVs is usually not investigated clearly [11][65].65. Conclusions
References
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Abels, E.R.; Maas, S.L.; Nieland, L.; Wei, Z.; Cheah, P.S.; Tai, E.; Kolsteeg, C.-J.; Dusoswa, S.A.; Ting, D.T.; Hickman, S.; et al. Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep. 2019, 28, 3105–3119.
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074.
- Shen, M.; Dong, C.; Ruan, X.; Yan, W.; Cao, M.; Pizzo, D.; Wu, X.; Yang, L.; Liu, L.; Ren, X.; et al. Chemotherapy-Induced Extracellular Vesicle miRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res. 2019, 79, 3608–3621.
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.
- Chen, S.; Tang, Y.; Liu, Y.; Zhang, P.; Lv, L.; Zhang, X.; Jia, L.; Zhou, Y. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 2019, 52, e12669.
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455.
- Zomer, A.; Maynard, C.; Verweij, F.; Kamermans, A.; Schäfer, R.; Beerling, E.; Schiffelers, R.; de Wit, E.; Berenguer, J.; Ellenbroek, S.; et al. In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell 2015, 161, 1046–1057.
- Ridder, K.; Keller, S.; Dams, M.; Rupp, A.-K.; Schlaudraff, J.; Del Turco, D.; Starmann, J.; Macas, J.; Karpova, D.; Devraj, K.; et al. Extracellular Vesicle-Mediated Transfer of Genetic Information between the Hematopoietic System and the Brain in Response to Inflammation. PLoS Biol. 2014, 12, e1001874.
- Ridder, K.; Sevko, A.; Heide, J.; Dams, M.; Rupp, A.-K.; Macas, J.; Starmann, J.; Tjwa, M.; Plate, K.H.; Sültmann, H.; et al. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. OncoImmunology 2015, 4, e1008371.
- Lu, J.J.; Yang, W.M.; Li, F.; Zhu, W.; Chen, Z. Tunneling Nanotubes Mediated microRNA-155 Intercellular Transportation Promotes Bladder Cancer Cells’ Invasive and Proliferative Capacity. Int. J. Nanomed. 2019, 14, 9731–9743.
- Ilahibaks, N.F.; Lei, Z.; Mol, E.A.; Deshantri, A.K.; Jiang, L.; Vader, P.; Sluijter, J.P.G. Biofabrication of cell-derived nanovesicles: A potential alternative to extracellular vesicles for regenerative medicine. Cells 2019, 8, 1509.
- Pucci, F.; Garris, C.; Lai, C.P.; Newton, A.; Pfirschke, C.; Engblom, C.; Alvarez, D.; Sprachman, M.; Evavold, C.; Magnuson, A.; et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science 2016, 352, 242–246.
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18.
- Friedman, R.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105.
- Zhang, B.; Yin, Y.; Lai, R.C.; Tan, S.S.; Choo, A.B.H.; Lim, S.K. Mesenchymal Stem Cells Secrete Immunologically Active Exosomes. Stem Cells Dev. 2014, 23, 1233–1244.
- Zheng, L.; Li, Z.; Ling, W.; Zhu, D.; Feng, Z.; Kong, L. Exosomes Derived from Dendritic Cells Attenuate Liver Injury by Modulating the Balance of Treg and Th17 Cells After Ischemia Reperfusion. Cell. Physiol. Biochem. 2018, 46, 740–756.
- Sverdlov, E.D. Amedeo Avogadro’s cry: What is 1 µg of exosomes? BioEssays 2012, 34, 873–875.
- Baglio, S.R.; Rooijers, K.; Koppers-Lalic, D.; Verweij, F.J.; Lanzón, M.P.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.M.; Baldini, N.; et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015, 6, 127.
- Chen, T.S.; Lai, R.C.; Lee, M.M.; Choo, A.B.H.; Lee, C.N.; Lim, S.K. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2009, 38, 215–224.
- Nolte-’t Hoen, E.N.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.M.; ’t Hoen Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285.
- Jenjaroenpun, P.; Kremenska, Y.; Nair, V.M.; Kremenskoy, M.; Joseph, B.; Kurochkin, I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013, 1, e201.
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS ONE 2015, 10, e0136133.
- Toh, W.S.; Lai, R.C.; Zhang, B.; Lim, S.K. MSC exosome works through a protein-based mechanism of action. Biochem. Soc. Trans. 2018, 46, 843–853.
- Borodavka, A.; Singaram, S.W.; Stockley, P.; Gelbart, W.M.; Ben-Shaul, A.; Tuma, R. Sizes of Long RNA Molecules Are Determined by the Branching Patterns of Their Secondary Structures. Biophys. J. 2016, 111, 2077–2085.
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information. Genes 2017, 8, 240.
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the Rna Content of the Exosomes Derived from Blood Serum and Urine and Its Potential as Biomarkers. Philos. Trans. R. Soc. Lond. Ser. Biol. Sci. 2014, 369, 20130502.
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.; Hughes, S.; Cheng, H.H.; Arroyo, J.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893.
- Arroyo, J.; Chevillet, J.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.; Bennett, C.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008.
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V.; Hessvik, N.P.; Phuyal, S.; Brech, A.; Sandvig, K.; et al. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1819, 1154–1163.
- Bryant, R.J.; Pawlowski, T.; Catto, J.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774.
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PLoS ONE 2014, 9, e92921.
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37.
- Miller, P.R.; Taylor, R.M.; Tran, B.Q.; Boyd, G.; Glaros, T.; Chavez, V.H.; Krishnakumar, R.; Sinha, A.; Poorey, K.; Williams, K.P.; et al. Extraction and biomolecular analysis of dermal interstitial fluid collected with hollow microneedles. Commun. Biol. 2018, 1, 173.
- Lindena, J.; Sommerfeld, U.; Höpfel, C.; Trautschold, I. Catalytic Enzyme Activity Concentration in Tissues of Man, Dog, Rabbit, Guinea Pig, Rat and Mouse. Approach to a Quantitative Diagnostic Enzymology, III. Communication. Clin. Chem. Lab. Med. 1986, 24.
- Wiklander, O.P.B.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316.
- Arraud, N.; Linares, R.; Tan, S.; Gounou, C.; Pasquet, J.-M.; Mornet, S.; Brisson, A.R. Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. J. Thromb. Haemost. 2014, 12, 614–627.
- Johnsen, K.B.; Gudbergsson, J.M.; Andresen, T.L.; Simonsen, J.B. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta (BBA) Bioenerg. 2019, 1871, 109–116.
- Li, Y.; He, X.; Li, Q.; Lai, H.; Zhang, H.; Hu, Z.; Li, Y.; Huang, S. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 2020, 18, 2851–2859.
- Matsumoto, A.; Takahashi, Y.; Chang, H.; Wu, Y.; Yamamoto, A.; Ishihama, Y.; Takakura, Y. Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. J. Extracell. Vesicles 2020, 9, 1696517.
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471.
- Tarr, L.; Oppenheimer, B.; Sager, R.V. The circulation time in various clinical conditions determined by the use of sodium dehydrocholate. Am. Heart J. 1933, 8, 766–786.
- Kandel, E.; Schwartz, J.; Jessel, T.M. Principles of Neural Science, 5th ed.; AccessNeurology; McGraw-Hill Medical: New York, NY, USA, 2013; ISBN 0838577016.
- Willekens, F.L.A.; Werre, J.M.; Kruijt, J.K.; Roerdinkholder-Stoelwinder, B.; Groenen-Döpp, Y.A.M.; Bos, A.G.V.D.; Bosman, G.J.C.G.M.; Van Berkel, T.J.C. Liver Kupffer cells rapidly remove red blood cell–derived vesicles from the circulation by scavenger receptors. Blood 2005, 105, 2141–2145.
- De Jong, O.G.; Murphy, D.E.; Mäger, I.; Willms, E.; Garcia-Guerra, A.; Gitz-Francois, J.J.; Lefferts, J.; Gupta, D.; Steenbeek, S.C.; Van Rheenen, J.; et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat. Commun. 2020, 11, 1113.
- Bonsergent, E.; Grisard, E.; Buchrieser, J.; Schwartz, O.; Théry, C.; Lavieu, G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 2021, 12, 1864.
- Joshi, B.S.; De Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455.
- Teo, S.L.Y.; Rennick, J.J.; Yuen, D.; Al-Wassiti, H.; Johnston, A.P.R.; Pouton, C.W. Unravelling cytosolic delivery of cell penetrating peptides with a quantitative endosomal escape assay. Nat. Commun. 2021, 12, 3721.
- Johannes, L.; Lucchino, M. Current Challenges in Delivery and Cytosolic Translocation of Therapeutic RNAs. Nucleic Acid Ther. 2018, 28, 178–193.
- Lagache, T.; Danos, O.; Holcman, D. Modeling the Step of Endosomal Escape during Cell Infection by a Nonenveloped Virus. Biophys. J. 2012, 102, 980–989.
- Shekari, F.; Nazari, A.; Kashani, S.A.; Hajizadeh-Saffar, E.; Lim, R.; Baharvand, H. Pre-clinical investigation of mesenchymal stromal cell-derived extracellular vesicles: A systematic review. Cytotherapy 2021, 23, 277–284.
- Wu, S.Y.; Singhania, A.; Burgess, M.; Putral, L.; Kirkpatrick, C.; Davies, N.; McMillan, N.A. Systemic delivery of E6/7 siRNA using novel lipidic particles and its application with cisplatin in cervical cancer mouse models. Gene Ther. 2010, 18, 14–22.
- Morrissey, D.V.; Blanchard, K.; Shaw, L.; Jensen, K.; Lockridge, J.A.; Dickinson, B.; McSwiggen, J.A.; Vargeese, C.; Bowman, K.; Shaffer, C.S.; et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 2005, 41, 1349–1356.
- Somiya, M. Where does the cargo go? Solutions to provide experimental support for the “extracellular vesicle cargo transfer hypothesis”. J. Cell Commun. Signal. 2020, 14, 135–146.
- Reshke, R.; Taylor, J.A.; Savard, A.; Guo, H.; Rhym, L.H.; Kowalski, P.; Trung, M.T.; Campbell, C.; Little, W.; Anderson, D.G.; et al. Reduction of the therapeutic dose of silencing RNA by packaging it in extracellular vesicles via a pre-microRNA backbone. Nat. Biomed. Eng. 2020, 4, 52–68.
- Eldh, M.; Lötvall, J.; Malmhäll, C.; Ekström, K. Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol. 2012, 50, 278–286.
- Meerson, A.; Ploug, T. Assessment of six commercial plasma small RNA isolation kits using qRT-PCR and electrophoretic separation: Higher recovery of microRNA following ultracentrifugation. Biol. Methods Protoc. 2016, 1, bpw003.
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10, 4333.
- Murphy, D.E.; de Jong, O.G.; Evers, M.J.W.; Nurazizah, M.; Schiffelers, R.M.; Vader, P. Natural or Synthetic RNA Delivery: A Stoichiometric Comparison of Extracellular Vesicles and Synthetic Nanoparticles. Nano Lett. 2021, 21, 1888–1895.
- Piffoux, M.; Silva, A.K.A.; Wilhelm, C.; Gazeau, F.; Tareste, D. Modification of Extracellular Vesicles by Fusion with Liposomes for the Design of Personalized Biogenic Drug Delivery Systems. ACS Nano 2018, 12, 6830–6842.
- Mao, Q.; Liang, X.-L.; Zhang, C.-L.; Pang, Y.-H.; Lu, Y.-X. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138-5p/Sirt1 axis. Stem Cell Res. Ther. 2019, 10, 1–14.
- Mendt, M.; Kamerkar, S.; Sugimoto, H.; McAndrews, K.M.; Wu, C.-C.; Gagea, M.; Yang, S.; Blanko, E.V.R.; Peng, Q.; Ma, X.; et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 2018, 3, e99263.
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345.
- Kooijmans, S.A.; Stremersch, S.; Braeckmans, K.; De Smedt, S.C.; Hendrix, A.; Wood, M.J.; Schiffelers, R.M.; Raemdonck, K.; Vader, P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release 2013, 172, 229–238.
- Piffoux, M.; Volatron, J.; Cherukula, K.; Aubertin, K.; Wilhelm, C.; Silva, A.K.; Gazeau, F. Engineering and loading therapeutic extracellular vesicles for clinical translation: A data reporting frame for comparability. Adv. Drug Deliv. Rev. 2021, 178, 113972.
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.L.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020, 159, 332–343.
- Fu, S.; Wang, Y.; Xia, X.; Zheng, J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261.
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013, 11, 88.
- Zhang, D.; Lee, H.; Zhu, Z.; Minhas, J.K.; Jin, Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Cell. Mol. Physiol. 2017, 312, L110–L121.
- Jeyaram, A.; Lamichhane, T.N.; Wang, S.; Zou, L.; Dahal, E.; Kronstadt, S.M.; Levy, D.; Parajuli, B.; Knudsen, D.; Chao, W.; et al. Enhanced Loading of Functional miRNA Cargo via pH Gradient Modification of Extracellular Vesicles. Mol. Ther. 2020, 28, 975–985.
- Ahn, S.Y.; Park, W.S.; Kim, Y.E.; Sung, D.K.; Sung, S.I.; Ahn, J.Y.; Chang, Y.S. Vascular endothelial growth factor mediates the therapeutic efficacy of mesenchymal stem cell-derived extracellular vesicles against neonatal hyperoxic lung injury. Exp. Mol. Med. 2018, 50, 1–12.
- Haghighitalab, A.; Matin, M.M.; Amin, A.; Minaee, S.; Bidkhori, H.R.; Doeppner, T.R.; Bahrami, A.R. Investigating the effects of IDO1, PTGS2, and TGF-β1 overexpression on immunomodulatory properties of hTERT-MSCs and their extracellular vesicles. Sci. Rep. 2021, 11, 7825.
- Eirin, A.; Zhu, X.-Y.; Puranik, A.S.; Tang, H.; McGurren, K.A.; van Wijnen, A.J.; Lerman, A.; Lerman, L.O. Mesenchymal stem cell–derived extracellular vesicles attenuate kidney inflammation. Kidney Int. 2017, 92, 114–124.
- Harting, M.T.; Srivastava, A.; Zhaorigetu, S.; Bair, H.; Prabhakara, K.S.; Furman, N.E.T.; Vykoukal, J.V.; Ruppert, K.A.; Cox, C.S.; Olson, S.D. Inflammation-Stimulated Mesenchymal Stromal Cell-Derived Extracellular Vesicles Attenuate Inflammation. Stem Cells 2018, 36, 79–90.
