Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Jinwei Zhang and Version 2 by Amina Yu.
Complex RNA–RNA interactions are increasingly known to play key roles in numerous biological processes from gene expression control to ribonucleoprotein granule formation. By contrast, the nature of these interactions and characteristics of their interfaces, especially those that involve partially or wholly structured RNAs, remain elusive. This entry describes different modalities of RNA–RNA interactions with an emphasis on those that depend on secondary, tertiary, or quaternary structure, and highlight a two-way relationship between RNA structure and interactions.
- RNA
- RNA structure
- RNA–RNA interactions
- tRNA
- T-box
- RNase P
1. Different Modalities of RNA–RNA Interactions: Components and Underlying Forces
The diverse ways RNAs can interact with each other are determined by the chemical, electrostatic, and geometric properties of their three components: the phosphodiester linkage, the ribose with its characteristic 2′-hydroxyl group, and the planar, heterocyclic nucleobase. While the latter, ribose- and nucleobase-mediated contacts are the primary drivers of RNA–RNA interactions, long-range interactions that involve the phosphodiester backbone are surprisingly common [1][4]. These generally involve OP1 or OP2 atoms as hydrogen bond acceptors and 2′-hydroxyl groups of an adjacently packed RNA segment as donors, forming the “ribose-phosphate zipper” motif [1][4]. It is possible that strong cationic ions such as Mg2+ or polyamines are needed to partially shield the charge of the phosphate backbone to allow such juxtaposition. Both the overall trajectory and local structure of the RNA phosphate backbone are more frequently recognized by basic amino acid residues of sequence-non-specific RNA-binding proteins. The ribose 2′-hydroxyl imparts significant structural rigidity over DNA strands and enforces A-form geometry in both double-stranded RNAs (dsRNA) and RNA–DNA hybrids. It also plays key roles in RNA recognition and ribozyme catalysis [2][5]. The versatile ribose 2′-hydroxyls can act as both donors and acceptors of hydrogen bonds, thereby allowing them to pair up and stitch together the backbone of adjacent antiparallel strands. These 2′-hydroxyl-2′-hydroxyl linkages, buttressed by neighboring 2′-hydroxyl contacts with purine N3 or pyrimidine O2 groups, form the prevalent “ribose zipper” motif, which contribute about ~2 kcal/mol of stabilizing energy [3][4][6,7].
The planar, aromatic nucleobases are the principal features that drive specific RNA–RNA interactions and can interact among themselves or with ribose 2′-hydroxyls and bridging and non-bridging oxygens of the phosphate backbone. Nucleobases can base pair or stack with other nucleobases and pack with ribose sugars via sugar-π interactions. Each nucleobase contains three geometric edges with distinct chemical compositions, namely the Watson–Crick, Hoogsteen, and sugar edges [5][8]. Nucleobases can further assume the anti (common) or syn (rare) configuration via the rotation of the glycosidic bond. Each edge can form base-pairing interactions with the same or a different edge in trans or in cis, thus producing a large variety of possible pairing configurations, most of which have been experimentally observed [5][8]. In addition to pairing interactions, whose vectors lie near perpendicular to the backbone trajectory, the aromatic nucleobases can stack along their backbone vector via π-π interactions. Importantly, if base-pairing interactions primarily create the local structures of RNA, stacking interactions principally control the global shape and trajectory of the RNA. This is exemplified by the double-helix shape of duplex nucleic acids, whose characteristic shape is largely a result of concatenated base stacking in the parallel displaced configuration rather than by base pairing. In addition, base stacking frequently mediates RNA–RNA interactions. Base intercalation is a key interaction that mediates the formation of the canonical tRNA elbow structure, where the G18 residue of D-loop inserts into the stacking gap of the T-loop [6][7][8][9,10,11], as well as the interdigitated double T-loop motif (IDTM) found in most T-box riboswitches and RNases P, which is formed by the reciprocal intercalation of two T-loops in head-to-tail opposition [9][10][12,13].
2. RNA Strand Assembly: From ssRNA to dsRNA, Triplex to G-Quadruplexes
All RNA transcripts start as ssRNA strands that still inherently repel each other from their negative charges. In this form, a balancing act between intra-strand stacking and repulsion from the neighboring phosphates controls the conformation of the ssRNA. As a result, polyA chains assume helical stacks due to strong stacking, while poorly stacked polyU chains form self-avoiding flexible polymers. The substantial hydrophobicity of the nucleobases motivates their approach towards each other against the electrostatic repulsion, which during their brief encounters explore ways to form base pairs and base stacks. For complementary ssRNA strands, once a few seed pairs form in the expected Watson–Crick geometry, the pairing propagates to lengthen the duplex in both directions. This pairing creates the basic building block of RNA secondary structure, in the form of a linear, antiparallel duplex.
Once formed, individual dsRNA segments are strongly inclined to stack coaxially with each other to form longer assemblies or “spines”. These end-to-end interactions are driven by favorable enthalpies from the π–π stacking between their termini base pairs, as well as favorable entropies associated with burying hydrophobic nucleobases that are otherwise exposed to the solvent. Such spontaneous coaxial assembly of dsRNA helices is exemplified by the tRNA folding process, during which the four helical segments stack coaxially into two longer stacks, before being joined at the elbow [11][12][13][14,15,16]. In the majority of RNA crystals, the helical segments are seen to form pseudo-infinite helices via end-to-end, coaxial stacking [14][17].
RNA duplexes generally exhibit A-form geometry characterized by deep, narrow major grooves and wide, shallow minor grooves. Both grooves can interact with an incoming ssRNA strand, sometimes at the same time with the same ssRNA, to form RNA triplexes. Notably, major groove triplexes that involve a third “Hoogsteen strand” are more stable than their minor groove counterparts [15][18], presumably due to the larger Hoogsteen edge in the major groove available for base triple formation than the sugar edge in the minor groove. First discovered by Gary Felsenfeld, David Davies, and Alexander Rich in 1957 in vitro [16][19], naturally occurring RNA triplexes have since then been identified in the telomerase RNA, spliceosome, Group II intron ribozymes, various riboswitches, and RNA stability elements such as the element for nuclear expression (ENE) [15][18]. Triplexes are also frequently found in RNA pseudoknots [17][20].
Beyond three-stranded RNAs, four strands of G-rich ssRNA can also interact among themselves to form G-quadruplexes. These extensively paired, highly stacked structures exhibit diverse topological arrangements, are more stable in RNA compared to DNA, and tend to fold slightly differently, favoring the parallel alignment in RNA form [18][19][21,22]. Serendipitously discovered by Martin Gellert, Marie N. Lipsett, and David R. Davies in 1962 [20][23], these robust and pervasive structures are stabilized by strong stacking interactions between adjacent G-quartet planes, which are quadrangular base pairs formed via tandem, circularizing interactions between their Watson–Crick and Hoogsteen edges. Interestingly, the large, hydrophobic quartet surfaces, if exposed to the solvent, can mediate robust intermolecular stacking interactions, as seen in the case of the dimeric, fluorogenic Corn RNA in the presence or absence of its chromophore ligand 3,5-difluoro-4-hydroxybenzylidene imidazolinone-2-oxime (DFHO) [21][24].
In summary, RNA–RNA interactions occur through diverse types of contacts (base pairing, base stacking, ribose zipper, A-minor, sugar-π interactions, etc.) and basic strand configurations (ssRNA, dsRNA, triplexes, and G-quadruplexes, etc.) More detailed analyses of the chemical structures of major RNA structural motifs such as K-turns, T-loops, A-minor motifs, and more, have been reviewed previously [22][23][24][25][26][25,26,27,28,29].
3. Interaction between ssRNAs: Base Pairing and Beyond
The most common type of RNA–RNA interactions are base-pairing interactions between complementary ssRNA strands. Prokaryotic small regulatory RNAs (sRNAs) target mRNAs in cis or in trans, primarily to repress gene expression by occluding the ribosome-binding sites, and secondarily by inducing RNase E cleavage via the dsRNA structure [27][28][30,31]. The establishment of such sRNA–mRNA pairing interactions frequently requires the action of RNA-binding chaperone proteins such as Hfq, ProQ, FinO, etc. (Figure 1a) [29][30][32,33]. Taking this protein-chaperoned RNA–RNA pairing one step further, the targeting ssRNA can first assemble into effector ribonucleoprotein (RNP) complexes before finding and annealing with the target mRNA. This mode of operation is exemplified by several RNA-targeted Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems including Type II (Cas9), Type III (Csm/Cmr), and Type VI (Cas13) CRISPRs [31][32][34,35]. Other types of sRNA may have more complex mechanisms of action, such as the 514-nt Staphylococcus aureus RNAIII. RNAIII, one of the largest sRNA identified so far, is proposed to repress the translation initiation of multiple mRNAs by annealing its CU-rich ssRNA loops with the complementary Shine–Dalgarno sequences of the mRNA [33][34][35][36,37,38].
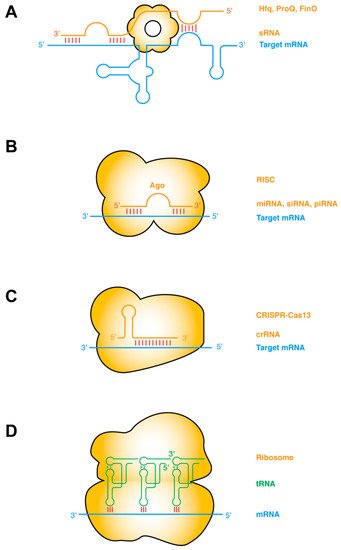
Figure 1. ssRNA–ssRNA interactions (A) Bacterial sRNAs form extensive base-pairing interactions with partially or wholly complementary segments of mRNAs, assisted by RNA chaperones such as Hfq, ProQ, FinO. sRNA binding remodels the mRNA structure to regulate its transcription, translation, stability, or decay, etc. (B) Eukaryotic small ssRNAs such as miRNAs, siRNAs, and piRNAs assemble with Ago proteins to form RNA-induced silencing complexes (RISC), and form perfect or imperfect base pairing with the target mRNA. Base pairing occurs first in the seed region and then in the supplementary pairing region. Such targeting leads to mRNA degradation or translation suppression. (C) RNA-targeting CRISPR-Cas13 binds the dsRNA duplex between the crispr RNA (crRNA) and the target mRNA, causing mRNA degradation and activating Cas13′s collateral RNase activity [31][34]. (D) Codon–anticodon base-pairing interactions between tRNA and mRNA are further stabilized by adjacent contacts from the rRNA and tRNA. Intermolecular base-pairing interactions are indicated by red sticks. Target mRNAs are shown in blue, sRNAs and other targeting or guiding RNAs in orange, and tRNAs in green. Proteins that facilitate the RNA interactions are shown as orange bodies.
In eukaryotes, there are at least three major categories of short ssRNAs that function analogously to the prokaryotic sRNAs and CRISPR RNA (crRNAs): the small interfering RNAs (siRNA), microRNAs (miRNA), and PIWI-interacting RNAs (piRNA) (Figure 1b,c). Each of these ssRNAs is processed from RNA hairpins or longer ssRNAs, and subsequently handed over to effector Argonaute (AGO) proteins, forming RNA-induced silencing complexes (RISC) of different flavors (siRISC, miRISC, and piRISC) [36][37][38][39,40,41]. These RNP complexes then scan for their target mRNAs and once found either catalyze their cleavage or repress their translation. Interestingly, at least with miRISC, the ~22-nt-long miRNA can sequentially form two discontinuous segments of base-pairing, first with the seed region (nt 2–7), and subsequently with the supplementary pairing region (nt 13–16). The latter is enabled by a conformational change triggered by the initial seed–mRNA pairing. Structural analyses revealed that the seed–target duplex minor groove is inspected by AGO to ensure Watson–Crick pairing and reject altered pairing geometry such as G•U wobble pairing [39][42].
A notable variation in ssRNA–ssRNA interactions occurs when both ssRNAs are hosted in structured hairpin stem loops. Such interactions can mediate long-range “kissing” interactions that bring far-flung RNA helices together to form compact structures (Figure 1d). Perhaps the most notable intermolecular ssRNA–ssRNA interactions aided by RNA structure are the codon–anticodon interactions between the mRNA and the tRNAs on the ribosome [40][43]. This interaction is driven by partial structure formation of both RNA partners. The mRNA bound by the ribosome assumes an extended conformation poised for the incoming tRNAs, which has evolved a particular anticodon stem loop structure that pre-stacks the anticodon trinucleotide in a helical ssRNA trajectory [40][43]. The short 3-bp mRNA–tRNA duplex is axially stabilized by adjacent tRNA and rRNA contacts, such as cross-strand stacking by the R37 residue of the tRNA on one side and the C1400 residue of the 16S rRNA (Escherichia coli numbering) on the other [41][42][43][44,45,46]. The codon–anticodon duplex is further laterally stabilized, and its Watson–Crick geometry enforced by the minor group interactions from G530, A1492, and A1493 of the rRNA [41][42][43][44,45,46]. Intriguingly, similar base-pairing, helix-capping by cross-strand stacking, and supplementary minor groove contacts also occur outside the ribosome between the tRNA and the T-box riboswitches [10][44][45][13,47,48].
