Recently, extracellular vesicles (EVs) and their contents have been revealed to play crucial roles in the intrinsic intercellular communications and have received extensive attention as next-generation biomarkers for diagnosis of diseases such as cancers. However, due to the structural nature of the EVs, the precise isolation and characterization are extremely challenging. To this end, tremendous efforts have been made to develop bionano sensors for the precise and sensitive characterization of EVs from a complex biologic fluid.
- biosensors
- extracellular vesicle
- nanoparticle
- nanotechnology
- optical sensor
- electrochemical sensor
1. Introduction
Recently, extracellular vesicles (EVs) (50–200 nm in diameter) have emerged as a new biomarker to diagnose disease progression and monitor treatment efficacy because of their vital role in cell–cell communication and molecular exchange [1][2][3]. These cell-derived membrane-enclosed vesicles are physically stable, abundant in biological fluids (e.g., >10
EVs/mL of blood) and carry cell-specific cargos that are characteristic of their cells of origin, including proteins and nucleic acids (e.g., micro RNA (miRNA), messenger RNA (mRNA) and DNA) [4][5][6][7]. Typically, extracellular vesicles are enriched with a transport or fusion protein (e.g., caveolin-1), tetraspanins (e.g., CD63, CD9, CD81) and heat-shock proteins (Hsp 60, Hsp 70 and Hsp 90) due to the endosomal formation of vesicles [8]. Most of the nucleic acids (approximately 76% of the total oligonucleotides) inside EVs are non-coding RNAs (i.e., miRNAs), which are involved in many different roles and functions of cells as post-transcriptional gene regulators [9][10]. Furthermore, because EVs are actively secreted in large quantities by cells, they are promising biomarker candidates for the early diagnosis of diseases and sub-sequential monitoring of the therapeutic response from liquid biopsies in a minimally invasive manner [11].
Despite such clinical potential, the reliable isolation and analyses of EVs from complex biologic fluids are enormously challenging because of the small size and low density of these biomolecules, thereby necessitating an extensive sample preparation technique before measurements. Although ultracentrifugation remains the gold standard for isolation of extracellular vesicles, it still requires expensive instruments, tedious steps and may introduce some impurities [5][12]. Meanwhile, conventional analytical methods, including western blotting, enzyme-linked immunosorbent assay (ELISA) and flow cytometry, have also been facing technical hurdles owing to the limited sample volume, low sensitivity, and/or requirement of specialized high-end equipment [13][14][15][16][17]. For example, the quantification of EVs by ELISA has a limit of approximately 10
in 100-μL of the sample [18]. To improve the efficiency and sensitivity of EV isolation from a complex biologic fluid, various nanotechnology-based biosensors have been developed [19][20][21].
2. Surface Plasmon Resonance-Based Analysis Methods for EV Detection
The SPR-based analytical method is an efficient platform for detecting and characterizing molecular interactions between biologic molecules [22,23,24,25,26]. This method monitors local refractive index changes generated by the resonant oscillation of stimulated electrons near the sensing surface (i.e., nanoscale metal materials) upon the binding of a ligand to a target molecule [27,28,29,30,31]. Owing to this simple and distinctive phenomenon, SPR-based biosensors have gained extensive attention to develop label-free and real-time biosensors with minimal sample preparation procedures [32,33,34,35,36]. Furthermore, as the dimension of EVs perfectly matches the sensing depth of SPR (200 nm), SPR-based biosensors are effective for quantifying EVs from complex biologic fluids [37,38,39,40]. In this section, recent studies related to SPR-based exosomal detection platforms are briefly introduced.The SPR-based analytical method is an efficient platform for detecting and characterizing molecular interactions between biologic molecules [22][23][24][25][26]. This method monitors local refractive index changes generated by the resonant oscillation of stimulated electrons near the sensing surface (i.e., nanoscale metal materials) upon the binding of a ligand to a target molecule [27][28][29][30][31]. Owing to this simple and distinctive phenomenon, SPR-based biosensors have gained extensive attention to develop label-free and real-time biosensors with minimal sample preparation procedures[32][33][34][35][36]]. Furthermore, as the dimension of EVs perfectly matches the sensing depth of SPR (200 nm), SPR-based biosensors are effective for quantifying EVs from complex biologic fluids [37][38][39][40]. In this section, recent studies related to SPR-based exosomal detection platforms are briefly introduced.
Qiu et al. utilized a nano-thick titanium nitride (TiN) film as an alternative plasmonic material in the SPR biosensing system for sensitive detection of EVs derived from malignant glioma cells (U251) (
Figure 1A) [41]. Based on the high affinity between the TiN film surface and biotin, the TiN film was functionalized by biotinylated anti-epidermal growth factor receptor (EGFR) variant-III (EGFR vIII) antibodies or anti-CD63 to capture U251-derived EVs. Both an in vitro cell culture condition and serum were tested to validate the performance of the biosensor in a real sample condition (i.e., complex biologic fluid). The developed biosensor had a limit of detection (LOD) of 4.29 × 10
−3-µg/mL for a general EV marker (CD63) and 2.75 × 10
−3-µg/mL for the glioma cell-specific marker, EGFR vIII. Compared with well-known plasmonic metal materials, including gold (Au), TiN even demonstrated better sensitivity, while having a tunable plasmonic property in the visible to near-infrared spectrum range as well.
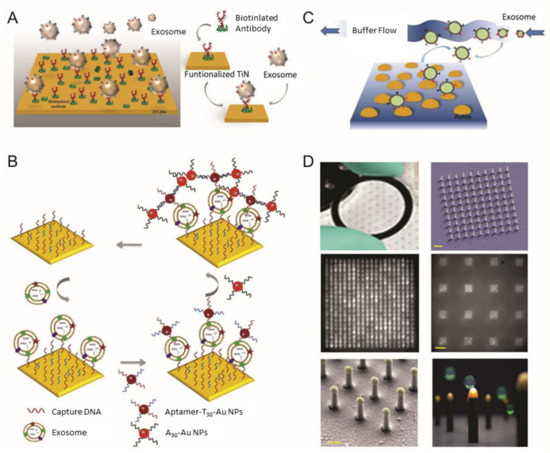
Surface plasmon resonance (SPR) biosensor for extracellular vesicles (EV) detection. (
) Schematic illustration of TiN nano-thick film functionalized by the anti-CD63 antibody for the detection of EVs; (
) dual Au Np-assisted signal amplification for the determination of EVs; (
) biophysical interaction of EV with self-assembled Au NIs without functionalization; (
) Au nanoplasmonic array functionalized with the anti-CD63 antibody for LSPR based digitalized detection of the EV. (reproduced with permission from [41] published by WILEY-VCH 2019, reproduced with permission from [42] published by Elsevier 2019, reproduced with permission from [43] published by Elsevier 2017 and reproduced with permission from [44] published by Public Library of Science 2018).
3. Colorimetric/Fluorescence-Based Analysis Methods for EV Detection
During the past few years, various types of nanomaterials with unique optical properties have been extensively applied to develop sensitive biosensors for EV analysis [21][45]. Among the optical techniques, a promising nanotechnology-based colorimetric biosensor has been established to detect exosomal biomarkers with the naked eye based on the extinction coefficient [46][47][48]. In general, nanotechnology-based colorimetric biosensors can be characterized into two groups based on their different properties of nanomaterials: catalytic properties (i.e., nanozymes) and inherent optical properties [49]. As an example of the catalytic property, Chen et al. improved the sensitivity of the traditional ELISA by fabricating a three-dimensional (3D) zinc oxide (ZnO) nanowires-coated scaffold chip device (
A) [50]. The presence of a hierarchical nanointerface, which was obtained by a 3D polydimethylsiloxane scaffold and free-standing ZnO nanowires, provides high capture efficiency of EVs as a result of the large surface area, as well as a size exclusion-like effect. After the isolation, a colorimetric assay based on horseradish peroxidase (HRP)-labeled antibody and 3,3′,5,5′-tetramethylbenzidine was performed for the quantitative analysis of EVs. Based on the improved sensitivity compared with a commercial ELISA, down to 2.2 × 10
EVs/μL was detected with a linear range of 2.2 × 10
–2.4 × 10
EVs/μL. Likewise, Zhang et al. integrated a microfluidic chip with self-assembled 3D herringbone nanopatterns to promote microscale mass transfer by increasing the surface area for efficient EV isolation [51]. Combined with a colorimetric method, the LOD achieved was down to 10 EVs/mL. Moreover, the quantitative detection of circulating exosomal CD24, EpCAM and folate receptor–alpha protein for diagnosing ovarian cancer was demonstrated using only 2 μL of plasma.
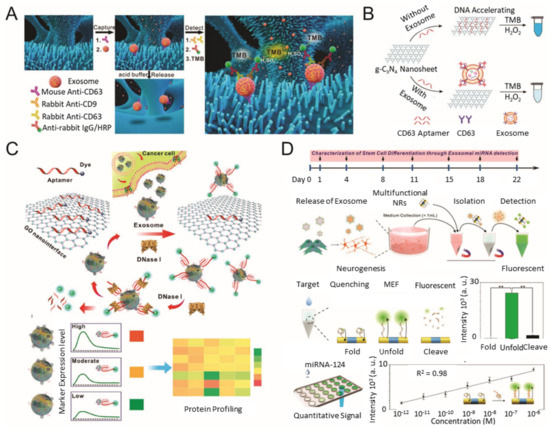
Colorimetric/fluorescence biosensor for EV detection. (
) Schematic diagram of the ZnO-nanowires-coated three-dimensional (3D) scaffold chip for EV detection; (
) utilization of peroxidase-like activity of graphitic carbon nitride nanosheets (g-C3N4 NSs); (
) fluorescence resonance energy transfer (FRET) biosensing system based on aptamer and graphene oxide (GO); (
) multifunctional nanorods (NRs) based exosomal miRNA signal amplification based on metal-enhanced fluorescence effect. (reproduced with permission from [50] published by Elsevier 2018, reproduced with permission from [52] published by American Chemical Society 2017, reproduced with permission from [53] published by American Chemical Society 2018 and reproduced with permission from [54] published by American Chemical Society 2019).
4. Conclusions and Future Perspective
This review article focused on biosensors that integrate nanotechnology with diverse analytical methods, including SPR-, fluorescence-, electrochemical- and Raman-based strategies for the effective measurement of EVs as biomarkers in diagnosing several diseases. Due to the inherent properties of nanomaterials (high surface-to-volume ratio, high reactivity and signal-enhancing effect), the precise and sensitive detection of EVs could be demonstrated. In addition, specific properties and multifunctionalities of each nanomaterial could facilitate the efficient measurement of EVs present in real samples. Most of the presented EV biosensors showed improved sensitivity by using functionalized nanomaterials and unique strategies. Besides the four representative analytical methods, others, such as field-effect-transistor [55][56], mechanical changes using a cantilever or quartz crystal microbalance [57][58][59] and giant magnetoresistance [60] could also be integrated with nanotechnologies to measure EVs. In this context, nanotechnology-integrated detection systems could be expanded to diverse analytical methods. In addition, label-free detection of EV can be developed by exploiting the high sensitivity of nanomaterials. One study presented a nanogap-integrated plasmonic sensing platform for EV detection without a Raman probe labeling step [61]. For improving the sensitivity, they used nanogap combining sub-volt dielectrophoretic trapping and Au NPs for real-time SERS imaging. Another study fabricated graphene-functionalized Au nanopyramids on SiO
and successfully characterized EVs isolated from different biologic sources using unbiased principal component analysis [62].
Besides detecting EVs by their specific surface proteins, encapsulated biomolecules, such as miRNA, mRNA and protein, could be important biomarkers for the diagnosis of diseases. Some exosomal miRNAs have been utilized as cancer biomarkers [63][64][65]. However, the expression levels of one miRNA could not indicate the presence of diseases precisely. Therefore, profiling of exosomal miRNA or other biomolecules will be more important for obtaining precise and personalized information, as well as diagnostic information or drug sensitivity. To this end, the isolation, rupture, and other pretreatment steps should be integrated into the sensing platform for collecting the encapsulated biomolecules efficiently. A powerful tool to leverage the functionality of biosensors is artificial intelligence-driven multi-technology bioprinting systems. On this wise, isolation, rupture, and other pretreatment steps should be integrated on the sensing platform for collecting the encapsulated biomolecules efficiently, as well as detection of the EV itself precisely. EV presents in diverse body fluids, such as blood, saliva, urine and interstitial fluids. For efficient measurement of EV in several real samples, preparation steps are very important to prevent pulse-positive signals and reduce trivial noise. Therefore, integrated platforms of sample preparation, detection, and analysis modules will be further developed with various nanotechnology to improve their performances. The concept of “organ-on-a-chip” is emerging for the etiology and drug screening of several diseases. For efficient drug testing, EV biosensors as a sensing module can provide real-time monitoring and fast response to the drugs on disease-emulated “organ-on-a-chip.” Currently, “organ-on-a-chip” operates as a proxy for in vitro and in silico research.
In conclusion, nanotechnology can be combined with EV biosensors to improve sensing capabilities, such as high selectivity, high sensitivity, straightforward and rapid detection, multi-detection and in situ monitoring. Here, we report the present status in the integration of nanotechnology-based biosensor platforms and analytical methods (SPR-, fluorescence-, electrochemical-based and Raman-based measurements) for detecting EVs. Each method has unique properties and suitable nanotechnologies are effortlessly utilized to improve the sensing performances. On this wise, isolation, rupture, and other pretreatment steps should be integrated on the sensing platform for collecting the encapsulated biomolecules efficiently, as well as detection of the EV itself precisely. EV presents in diverse body fluids, such as blood, saliva, urine and interstitial fluids. For efficient measurement of EV in several real samples, preparation steps are very important to prevent pulse-positive signals and reduce trivial noise. Therefore, integrated platforms of sample preparation, detection, and analysis modules will be further developed with various nanotechnology to improve their performances. In the immediate future, it is expected that micro total analysis systems and advanced “organ-on-a-chip” platforms with nanomaterial-based sensing modules for EVs will provide a robust in vitro drug development in platforms that can replace in vitro cell culture models and in vivo animal models and enable in vitro personalized investigations. Recently, multifunctional nanomaterials have been exploited to possess remarkable properties, such as enhancement of electron transfer reactions and improvement of quantum yield, for sensitive detection of optical properties. The development of nanotechnology will offer innovative and creative directions to develop novel biosensing platforms for EVs to increase the full recovery rate of diseases through the early diagnosis at a low level of exosomal biomarkers in real samples.
References
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorák, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856, doi:10.1038/sj.leu.2404132.
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta (BBA) Gen. Subj. 2012, 1820, 940–948, doi:10.1016/j.bbagen.2012.03.017.
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593, doi:10.1038/nri2567.
- Kang, Y.J.; Cutler, E.G.; Cho, H. Therapeutic nanoplatforms and delivery strategies for neurological disorders. Nano Converg. 2018, 5, 35, doi:10.1186/s40580-018-0168-8.
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950, doi:10.1021/acs.chemrev.7b00534.
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.-F.; Jacob, H.J.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 1–14, doi:10.1186/1471-2164-14-319.
- Johnsen, K.B.; Gudbergsson, J.M.; Andresen, T.L.; Simonsen, J.B. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta (BBA) Bioenerg. 2018, 1871, 109–116, doi:10.1016/j.bbcan.2018.11.006.
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638, doi:10.1038/s41571-018-0036-9.
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98, doi:10.1126/science.1090599.
- Lai, E.C. Micro RNAs are Complementary to 3′ UTR Sequence Motifs that Mediate Negative Post-Transcriptional Regulation. Nat. Genet. 2002, 30, 363.
- Xu, H.; Ye, B.-C. Advances in biosensing technologies for analysis of cancer-derived exosomes. TrAC Trends Anal. Chem. 2020, 123, 115773, doi:10.1016/j.trac.2019.115773.
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.-Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C.; et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017, 11, 6641–6651, doi:10.1021/acsnano.7b00549.
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. In Proteomic Profiling; Springer: Berlin/Heidelberg, Germany, 2015; pp. 179–209.
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernández, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182, doi:10.1038/nature14581.
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome Isolation for Proteomic Analyses and RNA Profiling. In Serum/Plasma Proteomics; Springer: Berlin/Heidelberg, Germany,2011; pp. 235–246.
- Gangoda, L.; Liem, M.; Ang, C.-S.; Keerthikumar, S.; Adda, C.G.; Parker, B.S.; Mathivanan, S. Proteomic Profiling of Exosomes Secreted by Breast Cancer Cells with Varying Metastatic Potential. Proteomics 2017, 17, 1600370, doi:10.1002/pmic.201600370.
- Yu, J.; Lin, Y.; Xiong, X.; Li, K.; Yao, Z.; Dong, H.; Jiang, Z.; Yu, D.; Yeung, S.-C.J.; Zhang, H. Detection of Exosomal PD-L1 RNA in Saliva of Patients With Periodontitis. Front. Genet. 2019, 10, 202, doi:10.3389/fgene.2019.00202.
- Merdalimova, A.; Chernyshev, V.; Nozdriukhin, D.; Rudakovskaya, P.; Gorin, D.A.; Yashchenok, A.M. Identification and Analysis of Exosomes by Surface-Enhanced Raman Spectroscopy. Appl. Sci. 2019, 9, 1135, doi:10.3390/app9061135.
- Bu, J.; Shim, J.-E.; Lee, T.H.; Cho, Y.-H. Multi-Modal liquid biopsy platform for cancer screening: Screening both cancer-associated rare cells and cancer cell-derived vesicles on the fabric filters for a reliable liquid biopsy analysis. Nano Converg. 2019, 6, 1–8, doi:10.1186/s40580-019-0204-3.
- Im, H.; Lee, K.; Weissleder, R.; Lee, H.; Castro, C.M. Novel nanosensing technologies for exosome detection and profiling. Lab Chip 2017, 17, 2892–2898, doi:10.1039/c7lc00247e.
- Shao, B.; Xiao, Z. Recent achievements in exosomal biomarkers detection by nanomaterials-based optical biosensors—A review. Anal. Chim. Acta 2020, 1114, 74–84, doi:10.1016/j.aca.2020.02.041.
- Singh, P. SPR Biosensors: Historical Perspectives and Current Challenges. Sens. Actuators B Chem. 2016, 229, 110–130, doi:10.1016/j.snb.2016.01.118.
- Kim, D.; Choi, E.; Lee, C.; Choi, Y.; Kim, H.; Yu, T.; Piao, Y. Highly sensitive and selective visual detection of Cr (VI) ions based on etching of silver-coated gold nanorods. Nano Converg. 2019, 6, 34–39, doi:10.1186/s40580-019-0206-1.
- Kim, D.W.; Chun, H.J.; Kim, J.-H.; Yoon, H.; Yoon, H.C. A non-spectroscopic optical biosensor for the detection of pathogenic Salmonella Typhimurium based on a stem-loop DNA probe and retro-reflective signaling. Nano Converg. 2019, 6, 16, doi:10.1186/s40580-019-0186-1.
- Ma, Y.; Dong, B.; Lee, C. Progress of infrared guided-wave nanophotonic sensors and devices. Nano Converg. 2020, 7, 12–34, doi:10.1186/s40580-020-00222-xs.
- Lee, J.-H.; Kim, B.-C.; Oh, B.-K.; Choi, J.-W. Highly sensitive localized surface plasmon resonance immunosensor for label-free detection of HIV-1. Nanomedicine 2013, 9, 1018–1026, doi:10.1016/j.nano.2013.03.005.
- Gong, C.; Leite, M. Noble Metal Alloys for Plasmonics. ACS Photonics 2016, 3, 507–513, doi:10.1021/acsphotonics.5b00586.
- Haes, A.J.; Zou, S.; Schatz, G.C.; Van Duyne, R.P. Nanoscale Optical Biosensor: Short Range Distance Dependence of the Localized Surface Plasmon Resonance of Noble Metal Nanoparticles. J. Phys. Chem. B 2004, 108, 6961–6968, doi:10.1021/jp036261n.
- Sugawa, K.; Tahara, H.; Yamashita, A.; Otsuki, J.; Sagara, T.; Harumoto, T.; Yanagida, S. Refractive Index Susceptibility of the Plasmonic Palladium Nanoparticle: Potential as the Third Plasmonic Sensing Material. ACS Nano 2015, 9, 1895–1904, doi:10.1021/nn506800a.
- Haes, A.J.; Zou, S.; Schatz, G.C.; Van Duyne, R.P. A Nanoscale Optical Biosensor: The Long Range Distance Dependence of the Localized Surface Plasmon Resonance of Noble Metal Nanoparticles. J. Phys. Chem. B 2004, 108, 109–116, doi:10.1021/jp0361327.
- Yang, L.; Lee, J.-H.; Rathnam, C.; Hou, Y.; Choi, J.-W.; Lee, K.-B. Dual-Enhanced Raman Scattering-Based Characterization of Stem Cell Differentiation Using Graphene-Plasmonic Hybrid Nanoarray. Nano Lett. 2019, 19, 8138–8148, doi:10.1021/acs.nanolett.9b03402.
- Jain, P.K.; Huang, X.; El-Sayed, I.H.; El-Sayed, M.A. Review of Some Interesting Surface Plasmon Resonance-enhanced Properties of Noble Metal Nanoparticles and Their Applications to Biosystems. Plasmonics 2007, 2, 107–118, doi:10.1007/s11468-007-9031-1.
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510, doi:10.3390/s150510481.
- Brolo, A.G. Plasmonics for future biosensors. Nat. Photonics 2012, 6, 709–713, doi:10.1038/nphoton.2012.266.
- Belotelov, V.; Akimov, I.; Pohl, M.; Kalish, A.; Kasture, S.; Vengurlekar, A.; Gopal, A.; Kotov, V.; Yakovlev, D.; Zvezdin, A. Intensity magnetooptical effect in magnetoplasmonic crystals. J. Phys. Conference Ser. 2011, 303, 012038.
- Rizal, C.; Belotelov, V. Sensitivity comparison of surface plasmon resonance (SPR) and magneto-optic SPR biosensors. Eur. Phys. J. Plus 2019, 134, 435, doi:10.1140/epjp/i2019-12819-3.
- Leung, A.; Shankar, P.; Mutharasan, R. A review of fiber-optic biosensors. Sens. Actuators B Chem. 2007, 125, 688–703, doi:10.1016/j.snb.2007.03.010.
- Jing, J.-Y.; Wang, Q.; Zhao, W.-M.; Wang, B.-T. Long-Range surface plasmon resonance and its sensing applications: A review. Opt. Lasers Eng. 2019, 112, 103–118, doi:10.1016/j.optlaseng.2018.09.013.
- Vlček, J.; Pištora, J.; Lesňák, M. Design of Plasmonic-Waveguiding Structures for Sensor Applications. Nanomaterials 2019, 9, 1227, doi:10.3390/nano9091227.
- Shpacovitch, V.; Hergenröder, R. Optical and surface plasmonic approaches to characterize extracellular vesicles. A review. Anal. Chim. Acta 2018, 1005, 1–15, doi:10.1016/j.aca.2017.11.066.
- Qiu, G.; Thakur, A.; Xu, C.; Ng, S.P.; Lee, Y.J.; Wu, C.M.L. Detection of Glioma-Derived Exosomes with the Biotinylated Antibody-Functionalized Titanium Nitride Plasmonic Biosensor. Adv. Funct. Mater. 2018, 29, 1806761, doi:10.1002/adfm.201806761.
- Wang, Q.; Zou, L.; Yang, X.; Liu, X.; Nie, W.; Zheng, Y.; Cheng, Q.; Wang, K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019, 135, 129–136, doi:10.1016/j.bios.2019.04.013.
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-Free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495, doi:10.1038/nbt.2886.
- Thakur, A.; Qiu, G.; Ng, S.P.; Guan, J.; Yue, J.; Lee, Y.J.; Wu, C.M.L. Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens. Bioelectron. 2017, 94, 400–407, doi:10.1016/j.bios.2017.03.036.
- Sun, J.; Lu, Y.; He, L.; Pang, J.; Yang, F.; Liu, Y. Colorimetric sensor array based on gold nanoparticles: Design principles and recent advances. TrAC Trends Anal. Chem. 2020, 122, 115754, doi:10.1016/j.trac.2019.115754.
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2018, 10, 18–33, doi:10.1039/C7NR06367A.
- Xia, Y.; Chen, T.; Chen, G.; Weng, Y.; Zeng, L.; Liao, Y.; Chen, W.; Lan, J.; Zhang, J.; Chen, J.-H. A nature-inspired colorimetric and fluorescent dual-modal biosensor for exosomes detection. Talanta 2020, 214, 120851, doi:10.1016/j.talanta.2020.120851.
- Cheng, N.; Du, D.; Wang, X.; Liu, N.; Xu, W.; Luo, Y.; Lin, Y. Recent Advances in Biosensors for Detecting Cancer-Derived Exosomes. Trends Biotechnol. 2019, 37, 1236–1254, doi:10.1016/j.tibtech.2019.04.008.
- Sabela, M.; Bechelany, M.; Janot, J.M.; Balme, S.; Bisetty, K. A Review of Gold and Silver Nanoparticle-Based Colorimetric Sensing Assays. Adv. Eng. Mater. 2017, 19, 1700270, doi:10.1002/adem.201700270.
- Chen, Z.; Cheng, S.-B.; Cao, P.; Qiu, Q.-F.; Chen, Y.; Xu, Y.; Xie, M.; Huang, W.-H. Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens. Bioelectron. 2018, 122, 211–216, doi:10.1016/j.bios.2018.09.033.
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.L.; Godwin, A.K.; Zeng, Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019, 3, 438–451, doi:10.1038/s41551-019-0356-9.
- Wang, Y.-M.; Liu, J.-W.; Adkins, G.; Shen, W.; Trinh, M.P.; Duan, L.-Y.; Jiang, J.-H.; Zhong, W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017, 89, 12327–12333, doi:10.1021/acs.analchem.7b03335.
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15, doi:10.1016/j.bios.2017.01.063.
- 63. Zhang, Q.; Wang, F.; Zhang, H.; Zhang, Y.; Liu, M.; Liu, Y. Universal Ti3C2 MXenes Based Self-Standard Ratiometric Fluorescence Resonance Energy Transfer Platform for Highly Sensitive Detection of exosomes. Anal. Chem. 2018, 90, 12737–12744, doi:10.1021/acs.analchem.8b03083.
- Tsang, D.K.H.; Lieberthal, T.; Watts, C.; Dunlop, I.E.; Ramadan, S.; Hernández, A.E.D.R.; Klein, N. Chemically Functionalised Graphene FET Biosensor for the Label-free Sensing of Exosomes. Sci. Rep. 2019, 9, 13946, doi:10.1038/s41598-019-50412-9.
- Yu, Y.; Li, Y.-T.; Jin, D.; Yang, F.; Wu, D.; Xiao, M.-M.; Zhang, H.; Zhang, Z.; Zhang, G.-J. Electrical and Label-Free Quantification of Exosomes with a Reduced Graphene Oxide Field Effect Transistor Biosensor. Anal. Chem. 2019, 91, 10679–10686, doi:10.1021/acs.analchem.9b01950.
- Etayash, H.; McGee, A.R.; Kaur, K.; Thundat, T. Nanomechanical sandwich assay for multiple cancer biomarkers in breast cancer cell-derived exosomes. Nanoscale 2016, 8, 15137–15141, doi:10.1039/C6NR03478K.
- Chang, Y.-J.; Yang, W.-T.; Wu, J.-C. Isolation and detection of exosomes via AAO membrane and QCM measurement. Microelectron. Eng. 2019, 216, 111094,, doi:10.1016/j.mee.2019.111094.
- Suthar, J.; Parsons, E.S.; Hoogenboom, B.W.; Williams, G.R.; Guldin, S. Acoustic Immunosensing of Exosomes Using a Quartz Crystal Microbalance with Dissipation Monitoring. Anal. Chem. 2020, 92, 4082–4093, doi:10.1021/acs.analchem.9b05736.
- Zhu, F.; Li, D.; Ding, Q.; Lei, C.; Ren, L.; Ding, X.; Sun, X. 2D magnetic MoS2–Fe3O4 hybrid nanostructures for ultrasensitive exosome detection in GMR sensor. Biosens. Bioelectron. 2020, 147, 111787, doi:10.1016/j.bios.2019.111787.
- Ertsgaard, C.T.; Wittenberg, N.J.; Klemme, D.J.; Barik, A.; Shih, W.-C.; Oh, S.-H. Integrated Nanogap Platform for Sub-Volt Dielectrophoretic Trapping and Real-Time Raman Imaging of Biological Nanoparticles. Nano Lett. 2018, 18, 5946–5953, doi:10.1021/acs.nanolett.8b02654.
- Yan, Z.; Dutta, S.; Liu, Z.; Yu, X.; Mesgarzadeh, N.; Ji, F.; Bitan, G.; Xie, Y.-H. A Label-Free Platform for Identification of Exosomes from Different Sources. ACS Sens. 2019, 4, 488–497, doi:10.1021/acssensors.8b01564.
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, U.D.; Kloecker, G.H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer 2009, 10, 42–46, doi:10.3816/clc.2009.n.006.
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PLoS ONE 2014, 9, e92921, doi:10.1371/journal.pone.0092921.
- Cho, S.; Yang, H.C.; Rhee, W.J. Simultaneous multiplexed detection of exosomal microRNAs and surface proteins for prostate cancer diagnosis. Biosens. Bioelectron. 2019, 146, 111749, doi:10.1016/j.bios.2019.111749.
