Light and brassinosteroid (BR) are external stimuli and internal cue respectively, that both play critical roles in a wide range of developmental and physiological process. Seedlings grown in the light exhibit photomorphogenesis, while BR promotes seedling etiolation. Light and BR oppositely control the development switch from skotomorphogenesis in the dark to photomorphogenesis in the light. Recent progress report that substantial components have been identified as hubs to integrate light and BR signals. Photomorphogenic repressors including COP1, PIFs, and AGB1 have been reported to elevate BR response, while photomorphogenesis-promoting factors such as HY5, BZS1, and NF-YCs have been proven to repress BR signal.
- photomorphogenesis
- skotomorphogenesis
- light
- brassinosteroids
- brassinolide
- Arabidopsis
1. Introduction
2. Signal Integration of Light and Brassinosteroid
Light and BR oppositely control the development switch from shotomorphogenesis in the dark to photomorphogenesis in the light. Numerous mutants involved in light and BR signals have overlapping phenotype, implicating the link between light and BR are intimate [4,5,119][4][5][20]. Genetic and biochemical evidences demonstrate that light signal share the highly component identity with BR signal to fine-tune plant growth and development. 2.1. Photomorphogenic Repressors Elevate Brassinosteroid Response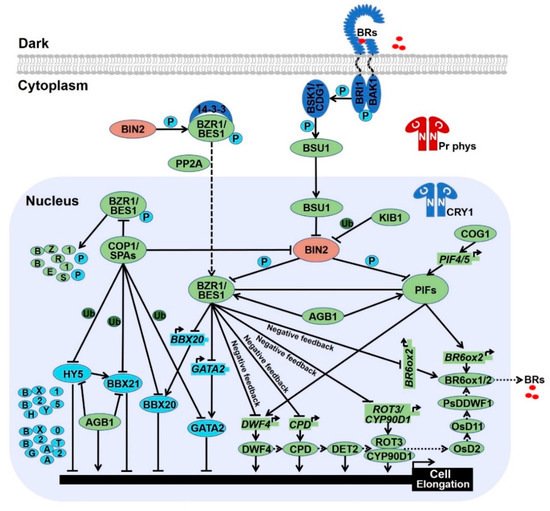
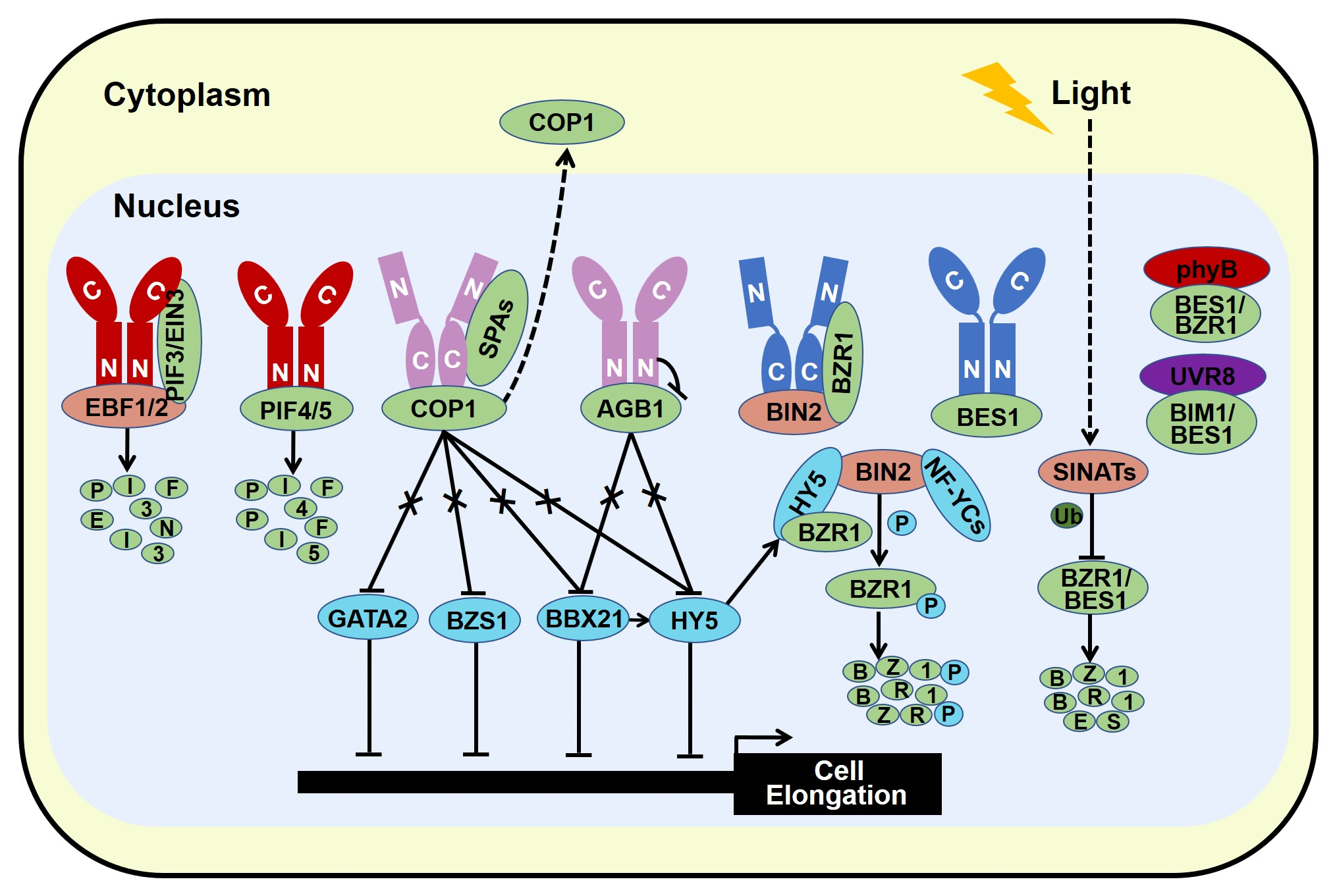
2.2. Photomorphogenesis-Promoting Factors Repress Brassinosteroid Signal
2.3. MicroRNAs Integrate Light and Brassinosteroid Signals
References
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230.
- Von Arnim, A.G.; Deng, X.W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 1994, 79, 1035–1045.
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-Regulated Plant Growth and Development. Curr. Top. Dev. Biol. 2010, 91, 29–66.
- Clouse, S.D.; Sasse, J.M. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu. Rev. Plant Biol. 1998, 49, 427–451.
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A Role for Brassinosteroids in Light-Dependent Development of Arabidopsis. Science 1996, 272, 398–401.
- Chaiwanon, J.; Wang, W.; Zhu, J.-Y.; Oh, E.; Wang, Z.-Y. Information Integration and Communication in Plant Growth Regulation. Cell 2016, 164, 1257–1268.
- Wang, Z.-Y.; Bai, M.-Y.; Oh, E.; Zhu, J.-Y. Brassinosteroid Signaling Network and Regulation of Photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724.
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809.
- Fan, X.-Y.; Sun, Y.; Cao, D.-M.; Bai, M.-Y.; Luo, X.-M.; Yang, H.-J.; Wei, C.-Q.; Zhu, S.-W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box Protein, Promotes Photomorphogenesis Downstream of Both Brassinosteroid and Light Signaling Pathways. Mol. Plant 2012, 5, 591–600.
- Bai, M.-Y.; Fan, M.; Oh, E.; Wang, Z.-Y. A Triple Helix-Loop-Helix/Basic Helix-Loop-Helix Cascade Controls Cell Elongation Downstream of Multiple Hormonal and Environmental Signaling Pathways in Arabidopsis. Plant Cell 2013, 24, 4917–4929.
- Bernardo-García, S.; de Lucas, M.; Martínez, C.; Espinosa-Ruiz, A.; Davière, J.-M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694.
- Liang, T.; Mei, S.; Shi, C.; Yang, Y.; Peng, Y.; Ma, L.; Wang, F.; Li, X.; Huang, X.; Yin, Y.; et al. UVR8 Interacts with BES1 and BIM1 to Regulate Transcription and Photomorphogenesis in Arabidopsis. Dev. Cell 2018, 44, 512–523.e5.
- Wang, W.; Lu, X.; Li, L.; Lian, H.; Mao, Z.; Xu, P.; Guo, T.; Xu, F.; Du, S.; Cao, X.; et al. Photoexcited CRYPTOCHROME1 Interacts with Dephosphorylated BES1 to Regulate Brassinosteroid Signaling and Photomorphogenesis in Arabidopsis. Plant Cell 2018, 30, 1989–2005.
- He, G.; Liu, J.; Dong, H.; Sun, J. The Blue-Light Receptor CRY1 Interacts with BZR1 and BIN2 to Modulate the Phosphorylation and Nuclear Function of BZR1 in Repressing BR Signaling in Arabidopsis. Mol. Plant 2019, 12, 689–703.
- Wu, J.; Wang, W.; Xu, P.; Pan, J.; Zhang, T.; Li, Y.; Li, G.; Yang, H.; Lian, H. phyB Interacts with BES1 to Regulate Brassinosteroid Signaling in Arabidopsis. Plant Cell Physiol. 2018, 60, 353–366.
- Dong, H.; Liu, J.; He, G.; Liu, P.; Sun, J. Photoexcited phytochrome B interacts with brassinazole resistant 1 to repress brassinosteroid signaling in Arabidopsis. J. Integr. Plant Biol. 2019, 62, 652–667.
- Li, J.; Terzaghi, W.; Gong, Y.; Li, C.; Ling, J.-J.; Fan, Y.; Qin, N.; Gong, X.; Zhu, D.; Deng, X.W. Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 2020, 11, 1592.
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; et al. Brassinosteroid-Activated BRI1-EMS-SUPPRESSOR 1 Inhibits Flavonoid Biosynthesis and Coordinates Growth and UV-B Stress Responses in Plants. Plant Cell 2020, 32, 3224–3239.
- Zhang, W.; Tang, Y.; Hu, Y.; Yang, Y.; Cai, J.; Liu, H.; Zhang, C.; Liu, X.; Hou, X. Arabidopsis NF-YCs play dual roles in repressing brassinosteroid biosynthesis and signaling during light-regulated hypocotyl elongation. Plant Cell 2021, 33, 2360–2374.
- Song, L.; Zhou, X.-Y.; Li, L.; Xue, L.-J.; Yang, X.; Xue, H.-W. Genome-Wide Analysis Revealed the Complex Regulatory Network of Brassinosteroid Effects in Photomorphogenesis. Mol. Plant 2009, 2, 755–772.
- Kang, J.-G.; Yun, J.; Kim, D.-H.; Chung, K.-S.; Fujioka, S.; Kim, J.-I.; Dae, H.-W.; Yoshida, S.; Takatsuto, S.; Song, P.-S.; et al. Light and Brassinosteroid Signals Are Integrated via a Dark-Induced Small G Protein in Etiolated Seedling Growth. Cell 2001, 105, 625–636.
- Park, D.H.; Lim, P.O.; Kim, J.S.; Cho, D.S.; Hong, S.H.; Gil Nam, H. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003, 34, 161–171.
- Wei, Z.; Yuan, T.; Tarkowská, D.; Kim, J.; Gil Nam, H.; Novak, O.; He, K.; Gou, X.; Li, J. Brassinosteroid Biosynthesis Is Modulated via a Transcription Factor Cascade of COG1, PIF4, and PIF5. Plant Physiol. 2017, 174, 1260–1273.
- Lease, K.A.; Wen, J.; Li, J.; Doke, J.T.; Liscum, E.; Walker, J.C. A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 2001, 13, 2631–2641.
- Jones, A.M.; Ecker, J.; Chen, J.-G. A Reevaluation of the Role of the Heterotrimeric G Protein in Coupling Light Responses in Arabidopsis. Plant Physiol. 2003, 131, 1623–1627.
- Xu, D.B.; Gao, S.Q.; Ma, Y.N.; Wang, X.T.; Feng, L.; Li, L.C.; Xu, Z.S.; Chen, Y.F.; Chen, M.; Ma, Y.Z. The G-Protein beta Subunit AGB1 Promotes Hypocotyl Elongation through Inhibiting Transcription Activation Function of BBX21 in Arabidopsis. Mol. Plant 2017, 10, 1206–1223.
- Lian, H.L.; Xu, P.B.; He, S.B.; Wu, J.; Pan, J.; Wang, W.X.; Xu, F.; Wang, S.; Pan, J.S.; Huang, J.R.; et al. Photoexcited CRYPTOCHROME 1 Interacts Directly with G-Protein beta Subunit AGB1 to Regulate the DNA-Binding Activity of HY5 and Photomorphogenesis in Arabidopsis. Mol. Plant 2018, 11, 1248–1263.
- Xu, P.; Lian, H.; Xu, F.; Zhang, T.; Wang, S.; Wang, W.; Du, S.; Huang, J.; Yang, H.-Q. Phytochrome B and AGB1 Coordinately Regulate Photomorphogenesis by Antagonistically Modulating PIF3 Stability in Arabidopsis. Mol. Plant 2019, 12, 229–247.
- Deng, X.W.; Matsui, M.; Wei, N.; Wagner, D.; Chu, A.M.; Feldmann, K.A.; Quail, P.H. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 1992, 71, 791–801.
- Zhang, T.; Xu, P.; Wang, W.; Wang, S.; Caruana, J.C.; Yang, H.Q.; Lian, H. Arabidopsis G-Protein beta Subunit AGB1 Interacts with BES1 to Regulate Brassinosteroid Signaling and Cell Elongation. Front. Plant Sci. 2017, 8, 2225.
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123.
- Ravindran, N.; Ramachandran, H.; Job, N.; Yadav, A.; Vaishak, K.; Datta, S. B-box protein BBX32 integrates light and brassinosteroid signals to inhibit cotyledon opening. Plant Physiol. 2021, 187, 446–461.
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.-Q.; Wang, X. Strigolactone/MAX2-Induced Degradation of Brassinosteroid Transcriptional Effector BES1 Regulates Shoot Branching. Dev. Cell 2013, 27, 681–688.
- Kim, E.J.; Lee, S.H.; Park, C.H.; Kim, S.H.; Hsu, C.C.; Xu, S.L.; Wang, Z.Y.; Kim, S.K.; Kim, T.W. Plant U-Box40 Mediates Degradation of the Brassinosteroid-Responsive Transcription Factor BZR1 in Arabidopsis Roots. Plant Cell 2019, 31, 791–808.
- Yang, M.; Li, C.; Cai, Z.; Hu, Y.; Nolan, T.; Yu, F.; Yin, Y.; Xie, Q.; Tang, G.; Wang, X. SINAT E3 Ligases Control the Light-Mediated Stability of the Brassinosteroid-Activated Transcription Factor BES1 in Arabidopsis. Dev. Cell 2017, 41, 47–58.e4.
- Kim, B.; Jeong, Y.J.; Corvalan, C.; Fujioka, S.; Cho, S.; Park, T.; Choe, S. Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 2014, 77, 737–747.
- Wang, L.; Tian, Y.; Shi, W.; Yu, P.; Hu, Y.; Lv, J.; Fu, C.; Fan, M.; Bai, M.-Y. The miR396-GRFs Module Mediates the Prevention of Photo-oxidative Damage by Brassinosteroids during Seedling De-Etiolation in Arabidopsis. Plant Cell 2020, 32, 2525–2542.
- Yang, Z.; Yan, B.; Dong, H.; He, G.; Zhou, Y.; Sun, J. BIC 1 acts as a transcriptional coactivator to promote brassinosteroid signaling and plant growth. EMBO J. 2021, 40, e104615.
- Ling, J.J.; Li, J.; Zhu, D.; Deng, X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA 2017, 114, 3539–3544.
- Ibañez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids Dominate Hormonal Regulation of Plant Thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e3.
- Li, Q.-F.; He, J.-X. BZR1 Interacts with HY5 to Mediate Brassinosteroid- and Light-Regulated Cotyledon Opening in Arabidopsis in Darkness. Mol. Plant 2016, 9, 113–125.
- Luo, X.-M.; Lin, W.-H.; Zhu, S.; Zhu, J.-Y.; Sun, Y.; Fan, X.-Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M.; et al. Integration of Light- and Brassinosteroid-Signaling Pathways by a GATA Transcription Factor in Arabidopsis. Dev. Cell 2010, 19, 872–883.
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.-H. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420.
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660.
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat Plants 2020, 6, 921–928.
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., 3rd; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792.
- Lin, L.-L.; Wu, C.-C.; Huang, H.-C.; Chen, H.-J.; Hsieh, H.-L.; Juan, H.-F. Identification of MicroRNA 395a in 24-Epibrassinolide-Regulated Root Growth of Arabidopsis thaliana Using MicroRNA Arrays. Int. J. Mol. Sci. 2013, 14, 14270–14286.
- Wang, T.; Zheng, Y.; Tang, Q.; Zhong, S.; Su, W.; Zheng, B. Brassinosteroids inhibit miRNA-mediated translational repression by decreasing AGO1 on the endoplasmic reticulum. J. Integr. Plant. Biol. 2021, 63, 1475–1490.
- Xia, K.; Ou, X.; Tang, H.; Wang, R.; Wu, P.; Jia, Y.; Wei, X.; Xu, X.; Kang, S.H.; Kim, S.K.; et al. Rice microRNA osa-miR1848 targets the obtusifoliol 14alpha-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 2015, 208, 790–802.
- Lau, K.; Podolec, R.; Chappuis, R.; Ulm, R.; Hothorn, M. Plant photoreceptors and their signaling components compete for COP 1 binding via VP peptide motifs. EMBO J. 2019, 38, e102140.
- Song, Z.Q.; Heng, Y.Q.; Bian, Y.T.; Xiao, Y.T.; Liu, J.J.; Zhao, X.H.; Jiang, Y.; Deng, X.W.; Xu, D.Q. BBX11 promotes red light-mediated photomorphogenic development by modulating phyB-PIF4 signaling. aBIOTECH 2021, 2, 117–130.
