You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Vicky Zhou and Version 1 by Meric Ozturk.
Being the predominant cause of disability, neurological diseases have received much attention from the global health community. Over a billion people suffer from one of the following neurological disorders: dementia, epilepsy, stroke, migraine, meningitis, Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, amyotrophic lateral sclerosis, Huntington’s disease, prion disease, or brain tumors. The diagnosis and treatment options are limited for many of these diseases. Aptamers, being small and non-immunogenic nucleic acid molecules that are easy to chemically modify, offer potential diagnostic and theragnostic applications to meet these needs.
- aptamer
- neuroscience
- neurological diseases
- neurological disorders
- neurotoxins
1. Introduction
Aptamers are single-stranded DNA, RNA, or synthetic XNA molecules that can fold into unique three-dimensional (3D) structures by which they bind their target molecules with high specificity and affinity [1,2][1][2]. An approach to selecting aptamers was achieved in 1990 by three separate groups [3,4,5][3][4][5]. The method is now known as the Systematic Evolution of Ligands by Exponential Enrichment (SELEX). Since then, many aptamers have been selected for use in areas of basic science and an increasing number of aptamers are under development for applications in therapeutics, diagnostics, and imaging [6,7,8,9,10][6][7][8][9][10].
The SELEX method simulates Darwinian evolution in vitro. It includes a number of selection rounds, and, after each round, exponential amplification of the “fittest” oligonucleotides (oligos) is carried out. One round of traditional SELEX includes three steps in the order (1) incubate the desired target molecule with a pool of oligos with a central 20–60 nucleotide region of randomized sequence surrounded by terminal regions of constant sequence that will be templates for later polymerase chain reactions (PCRs), (2) capture the oligos in the pool that successfully bind the target, and (3) amplify the captured oligos by either PCR or reverse transcription-PCR (rt-PCR), depending on the type of oligo, DNA or RNA respectively. Rounds of counter-selection are also included, in which the nucleic acid pool is passed over an empty supporting matrix, without the desired target or over the supporting matrix with one or more other molecules that might be structurally related to the target molecule. In counter selection, the oligos that do not bind the supporting matrix, or the structurally related molecules are captured and continued through more rounds of SELEX. Counter selection is performed for many purposes, such as to eliminate oligos that interact with (1) the supporting matrix, (2) analogs or molecules that are structurally related to the target, or (3) molecules present at high concentrations in the biological matrix (such as serum or tissue extract) in which the selected aptamer will function. The main goal of counter selection is to increase aptamer specificity for the desired target over other potential molecular competitors. Besides conventional SELEX, recently developed structure-switching SELEX is becoming popular [11,12,13][11][12][13]. In this approach, oligos are required to change their structures when they interact with the target, in order to dissociate from a complementary capture sequence. This ensures that the selected aptamers change in structure upon ligand binding. Other alternative SELEX methods have been developed including CE-SELEX, Cell-SELEX, and in-vivo SELEX, as summarized in Table 1. It is quite possible to combine different SELEX procedures to develop even more complex techniques. After aptamers are selected by SELEX, they are further modified to improve their specificities and affinities by a number of approaches that, in toto, can be referred to as maturation.
Table 1.
Summary of Alternative SELEX methods.
| SELEX Type | Features | Reference | |||
|---|---|---|---|---|---|
| Modification | Reference | ||||
| PhotoSELEX | Light sensitive oligonucleotides are excited by UV and covalently link to their target molecules | [14] | |||
| increases stability and resistance to 3′ exonuclease | 3′-3′ and 5′-5′ internucleotide linkage | [33] | |||
| Cell-SELEX | Whole cells are used for the selection of aptamers that bind cell surface targets | [15] | |||
| resistance to 3′ exonuclease | 3′ Biotin Conjugates | [34] | In vivo SELEX | Aptamers are selected from an oligonucleotide pool in living animals | [16] |
| increases nuclease resistance | 2′-fluoro (2′-F) Substitution | [35] | In silico SELEX | Computer programs are used to predict tertiary structure, affinity, and target interaction of aptamer candidates | [17] |
| 2′-amino (2′NH2) Substitution | [35] | CE-SELEX | Capillary electrophoresis is used to select high-affinity aptamers, which reduces the selection process time from weeks to days | ||
| 2′-O-methly (2′-OMe) Substitution | [36] | [18] | |||
| Spiegelmer Technology | After selection, aptamers are synthesized as unnatural L-oligonucleotides, which are more stable than D-oligonucleotides | [19] | |||
| Triazole replacement | [37] | Structure Switching SELEX | The nucleic acid pool has a short, unvaried sequence by which all oligonucleotides can be captured on a complementary sequence. The oligonucleotides are released when they switch structures to bind their target molecule. | [13] | |
| L-DNA | [38] | Magnetic-assisted Rapid Aptamer Selection (MARAS) | Magnetic nanoparticle-attached targets are used to capture aptamers in the presence of an externally applied rotating magnetic field with varying frequencies that influence the selected aptamer affinities. | [20] | |
| increases DNA nuclease resistance, destabilizes quadruplexes in aptamer structure | thiophosphoryl modifications | [39] | Artificially Expanded Genetic Information (AEGIS)-SELEX | The AEGIS-SELEX library is composed of oligonucleotides containing natural and non-natural nucleosides. These libraries have higher sequence diversities than libraries of oligonucleotides containing only natural nucleosides. | [21] |
| resistance to renal clearance | 5′-End with Cholesterol | [40] | Robotic Assisted-SELEX | Robotic platforms perform the selection without any manual intervention. It reduces the selection process to less than 2 days | [22] |
| 5′-End with Dialkyl Lipids | [41] | RAPID-SELEX | |||
| 5′-End with PEGylation | A conventional SELEX protocol, but without amplification. After each round, Kd values are measured, and the enriched aptamers are sent to HTS. | [42[23] | |||
| GO-SELEX | A conventional SELEX protocol with unbound oligos adsorbed by graphene oxide (GO) | [24] | |||
| Sol-gel SELEX | The desired aptamer target is immobilized on a microfluidic device | [25] | |||
| Conditional SELEX | This method enables the selection of aptamers that only function under the chosen condition such as when they are in the presence of a regulatory molecule | [26] | |||
| Tailored SELEX | The library sequences do not have primer complements and SELEX is performed in the absence of primer complements. To amplify the selected oligonucleotides, the primer complements are ligated with primers. This method prevents the primer complements on the oligonucleotides from being part of the selected aptamer structure that binds to target | [19] | |||
| SPR-SELEX | The desired target is immobilized on an SPR chip and the oligo pool injected on the biosensor chip for aptamer selection. | [27] | |||
| Chimeric SELEX | Two or more libraries are used to isolate functionally different aptamers, which are then fused to create a dual function aptamer. | [28] | |||
| FRELEX | Random 8mers are used to capture the aptamers in Phase I and the target molecule is free in solution during Phase II of selection. This method allows for a true free aptamer selection strategy. | [29] |
There are several considerations in selecting and maturing aptamers. First, the structures of aptamers and aptamer-target interactions are affected by the temperature and the buffer components, including ions, ionic strength, and pH. Thus, one of the most important points in SELEX design is to mimic the environment (biological matrix or salt/ buffer composition) during selection in which the aptamer molecular target is found and the interaction between aptamer and target will occur. Choosing appropriate buffer compositions and incubation temperatures will ensure the optimal performance of the selected aptamers under the conditions in which they will be applied [4,30,31,32][4][30][31][32]. Second, aptamer stability may need to be improved, as DNA and RNA aptamers are potential targets for nuclease attack. This is especially the case for RNA, because of the 2′OH group, which is used by ribonucleases in an electrophilic attack of the phosphate of the nucleic acid backbone for hydrolysis. Thus, RNA is more labile to high temperatures and pH than DNA. Aptamers can be matured by post-selection chemical modifications to overcome this susceptibility. However, such modifications can result in the aptamer losing specificity and/or affinity. Therefore, it is usually better to include modified nucleotides during SELEX. Many possible aptamer modifications are listed in Table 2.
Table 2.
Possible Aptamer Modifications with their Characteristics.
| Feature | |
|---|---|
| ] | |
| improving binding affinity and target selectivity | |
| base modifications (SOMAmers) | |
| [ | |
| 43 | |
| ] | |
| structure-based modifications | [44] |
Aptamers can be selected that selectively bind most viruses, cells, bacteria, proteins, toxins, and peptides with high affinities [45,46,47,48][45][46][47][48]. They can be used several times without noticeable disruption of activity. In other words, they can be separated from their targets for further use. By contrast, antibodies can only be utilized only a few times before they lose functionality. Nucleic acid aptamers can also be matured to be more stable than antibodies and enzymes, particularly at higher temperatures [2]. Moreover, once the sequence of an aptamer is known, its chemical synthesis and purification to homogeneity are highly reproducible and inexpensive. These properties give aptamers the potential to substitute for antibodies as components of sensing units [49]. The flexibility of aptamers’ structures and their ability to “mask” regions of an aptamer sequence with complementary oligos provides options for signaling that allow the incorporation of aptamers into many sensor platforms, which are not readily adaptable to antibodies. These properties motivate aptamer-based biosensor development [50,51,52][50][51][52].
Developing new tools for diagnosis is one of the most salient areas in the biosensor field, with many investigators seeking to create easy, efficient, and cheaper methods to achieve early diagnosis and precision medicine. The inclusion of aptamers in biosensors (named “aptasensors”) took root in the 1990s. An early example of aptamers as bio-recognition elements is an optical biosensor that used fluorescently labeled aptamers against human neutrophil elastase in a homogenous assay [53]. Since then, many types of aptasensor designs have been reported, most with aptamers on solid supports, and with electrochemical, optical, mechanical, and acoustic signals [45,54,55,56][45][54][55][56]. Some examples are discussed later in this article.
Aptamers have also been applied in therapeutics. Modifications that result in increased half-lives and improved pharmacokinetics make some aptamers good options for clinical application. Compared with antibodies, aptamers are much less immunogenic, and their actions can be inhibited by reverse complementary oligonucleotides (Munzar et al., 2019). These important characteristics recommend aptamers for development as therapeutics. Macugen (pegaptanib sodium), which selectively recognizes vascular endothelial growth factor (VEGF), was the first aptamer to be approved as a therapeutic agent (Tobin, 2006). Since then, several aptamers have reached different stages of clinical trials in which they are being tested for treating medical conditions like small cell lung cancer, von Willebrand factor-related disorder, angiomas, and renal cell carcinomas [51,57,58,59,60][51][57][58][59][60].
Another potential therapeutic application of aptamers involves delivery systems [61,62][61][62]. To achieve efficient results in these systems, aptamers should recognize cell surface proteins in their native forms. Cell-SELEX was developed to select against cell-surface proteins that are frequently difficult to purify in their native forms [63]. An aptamer that recognizes a cell surface protein can coopt the receptor to internalize a cargo attached to the aptamer. With this method, one can select aptamers to receptors primarily expressed on a single cell type that can be targeted in vivo [57]. For instance, anti-PSMA (prostate-specific membrane antigen) aptamers were developed for the delivery of siRNAs to tumors in mice or cultured cells [62,64,65,66][62][64][65][66].
In neuroscience, the use of aptamers has, so far, been limited, albeit while having a very wide range of potential applications. Aptamers have been reported as possible treatment options for neurodegenerative diseases that result in the loss of central nervous system (CNS) structure and function, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Creutzfeldt–Jakob disease, motor neuron diseases, and Huntington’s disease (HD) (Figure 1). Currently, around 50 million people in the world face dementia, for which the global healthcare cost is close to a trillion US dollars per year. This makes dementias one of the biggest medical and economic problems for our society [67]. As well, according to the United Nations and the World Health Organization, the world population over 65 years old is estimated to double by 2050. Therefore, age-dependent neurodegenerative diseases will require increasing expenditures for diagnosis and treatment [67]. These critical demands for methods to facilitate early diagnosis and new therapeutic applications could be met by aptamers, which could also be solutions for some of the underlying complications [68,69][68][69].
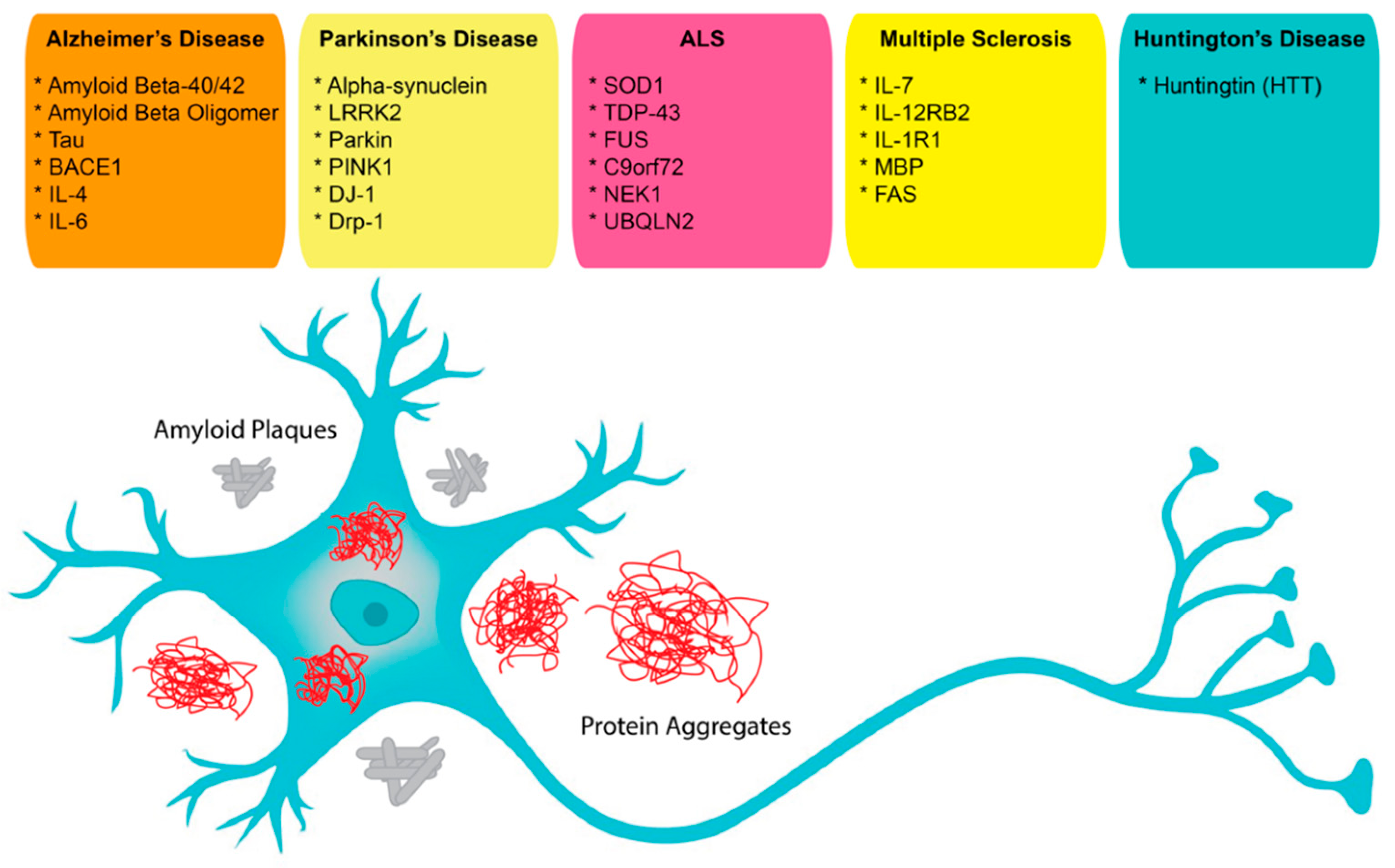
Figure 1. Possible aptamer targets in neuroscience.
Aptamers are being developed for applications in brain imaging technologies (MRI, PET, etc.), neurotransmitter visualization, diagnosis, and therapeutics for brain tumors and other brain-related diseases. In many of these approaches, aptamers are labeled with radiotracers, such as fluorine-18 for PET imaging [70].
2. Diagnostic and Therapeutic Applications
Neurodegenerative diseases (NDs) involve the degeneration of neurons in the brain that results in the loss of structures and functions of the central nervous system. ND can be caused by aging, genetic and environmental factors. Dementia is one of the most common age-related NDs as is Alzheimer’s disease (AD). Parkinson’s disease (PD) can be due to genetic mutations and/or environmental toxins. Amyotrophic lateral sclerosis (ALS), Multiple Sclerosis (MS), Huntington’s disease (HD), and prion diseases are other common NDs with genetic links. Unfortunately, neither effective cures nor early detection strategies are available for these diseases. Like the widely used antibodies, aptamers have become attractive agents to apply to developing novel biosensors for early diagnosis of NDs and cure of these diseases [88][71].2.1. Alzheimer’s Disease
As an age-related progressive brain disorder resulting in mental deficiencies, AD is the most common form of dementia. Its pathology is characterized by the aggregation of amyloid-β (Aβ) derived from the amyloid precursor protein (APP) and initiated in the brain region of the hippocampus. Although scientists hypothesized that Aβ-induced neurotoxicity was correlated with insoluble Aβ plaques (AβP) and fibrils (AβF), recent evidence indicates that soluble Aβ oligomers (AβO) are also associated with AD onset. Therefore, AβO has been identified as an attractive biomarker for early diagnosis of AD, and Aβ and tau are considered as significant therapeutic targets for treating AD [89][72].
2.2. Parkinson’s Disease
Parkinson’s disease (PD) is the second most common neurodegenerative disease, affecting about seven million people globally. While being a progressive disease characterized by motor and nonmotor features, it has significant clinical impacts on patients due to their loss of mobility and muscle control. Loss of striatal dopaminergic neurons and neuronal loss in nondopaminergic areas characterize PD. Loss of neurons is generally associated with characteristic Lewy body formation. Thus, the presence of Lewy bodies containing α-syn oligomers in the brain is considered a possible therapeutic and diagnostic target for PD.2.3. Multiple Sclerosis
Multiple sclerosis (MS) is an autoimmune disease in which the myelin sheaths surrounding the neuronal axons in the brain and spinal cord are destroyed. Disease progression is highly unpredictable and refractive to treatment at later stages. Characteristic pathologies of the disease are abnormal activation of microglia and astrocytes, which results in oligodendrocytes and neuronal cell degeneration. While there is no effective cure or diagnosis of the disease, the number of MS patients in the world is greater than 2.5 million [109][73]. Specific biomarkers that could identify MS early in its progression are not available, anti-inflammatory therapies have faced failures and they only prevent relapses.2.4. Amyotrophic Lateral Sclerosis
Dysregulation or alteration of receptor expression levels have been demonstrated in various diseases. Abnormally activated AMPA receptors, which are associated with amyotrophic lateral sclerosis (ALS), are potential drug candidates for ALS treatment. RNA aptamer AN58, raised against the GluR2Q flip AMPA receptor, competitively inhibited the receptor. Its nanomolar affinity is better than NBQX, one of the current best competitive inhibitors. AN58 demonstrated the highest affinity towards GluR2 and higher selectivity for the GluR4 among all AMPA receptor subunits. In short, AN58 is a potential inhibitor with nM affinity of the GluR2 AMPA receptors [116][74].
Another characteristic of ALS pathology is the toxic accumulation of the TAR DNA-binding protein 43 (TDP-43). Interaction of TDP-43 with RNA results in the regulation of RNA transcription, splicing, transport, and translation. Zacco et al. demonstrated that native partners of TDP-43 can be used to inhibit its aggregation. This inhibition occurs in a length-dependent manner, such that shorter oligos interfere with the aggregation better than long ones. Their study indicates the possibility of increasing protein solubility using a natural interaction that can be adapted as a new therapeutic approach [117][75].
2.5. Huntington Disease
Huntington’s disease (HD) is an incurable hereditary ND, impairing motor and cognitive functions. Huntingtin (HTT) protein is essential for neuronal development; however, some mutations lead to the development of HD pathology. Increases in the number of CAG repeats result in elongation of a polyglutamine stretch of HTT and convert it to a toxic protein, which causes HD. Similar to AD and PD, inhibition of protein aggregation is a promising strategy to slow or stop HD progression [118][76]. As mentioned before, aptamers, being low- to nonimmunogenic and nontoxic, emerge as robust candidates to interfere with protein–protein interaction and inhibit toxic protein aggregation in such diseases.
High-affinity RNA aptamers were selected against monomeric mHTT (51Q-HTT) and shown to effectively inhibit its aggregation in vitro. Such inhibition diminished oxidative stress in red blood cells (RBCs) and is associated with reduced leakage of a thioflavin-induced fluorescence from liposomes. The presence of aptamers rescued an endocytotic defect and blocked sequestration of glyceraldehyde-3-phosphate dehydrogenase by aggregated mHTT in a yeast model of Huntington’s. Some of these aptamers did not recognize the nonpathogenic, 20Q-HTT, and some increased the number of mHTT in the soluble fraction of yeast. When coexpressed, two successful aptamers increased the efficiency of inhibiting aggregation and improved cell survival. This study implies that using aptamers might be a viable strategy to slow the course of HD [119][77].
The long polygutamyl stretches cause slight changes in the HTT 3D structure and changes its activity. Indirect modulation of the affected structure using this elongated region on the protein might provide an alternative approach to treatment. G-quadruplex forming DNA aptamers (MS1 to MS4) that bind mHTT significantly decreased the activation by mHTT of the basal histone H3 lysine 27 trimethylation (H3K27me3) activity of the polycomb repressive complex 2 (PRC2) in neuronal progenitor cells (NPCs) from an individual with HD, but not in NPCs from a healthy individual [120][78]. With this study, DNA aptamers were successfully applied to preferentially target mHTT and to modulate its activity. These two examples provide novel structure-based approaches to effective treatments of mHTT toxicity.
2.6. Prion Disease
Transmissible spongiform encephalopathies (TSEs) are a type of neurodegenerative disorder that affect mammals. The pathologic agent associated with these diseases is a misfolded prion protein (PrP). The molecular mechanism (s) resulting in the structural changes that covert the cellular PrP (PrPC) to a pathogenic conformer (PrPSc) are only partially understood, with the most common hypothesis being that a molecular cofactor, acting as a catalyst, favors the transition from PrPC to PrPSc [121][79].
Three G-quadruplex-forming aptamer sequences were identified for different forms of PrP [122][80]. These quadruplexes were reported to have high affinity and specificity toward PrP (Kd: 62 nM–630 nM) and a weak affinity for the PrP-β oligomer that mimics the early stage of PrPSc formation. By various analyses, including ITC, SPR, and CD spectroscopy, a high-affinity binding for PrP was associated with a quadruplex structure and PrP terminal domains were required for aptamer binding. This study also provided evidence for a mutual unwinding of nucleic acid and protein upon their interaction. The quadruplex unwinding-activity of PrP was carried out by the intrinsically unstructured N-terminal domain and the DNAs promoted the unfolding of the PrP structured C-terminal domain.
2.7. Brain Tumors
Brain tumors are among the most fatal forms of cancer. For example, about two-thirds of adults diagnosed with glioblastoma (a type of brain cancer) lose their life within two years. Brain tumors are also the most common and lethal of all pediatric solid tumors. Children who survive with these tumors also often suffer from the long-term consequences of the necessary medical interventions, such as surgeries and chemotherapies.
Gliomas (glioblastoma, ependymomas, astrocytomas, and oligodendrogliomas) make up almost 80% of all malignant primary tumors of the brain. Glioblastoma multiforme (GBM) is the most frequently observed type of primary astrocytoma constituting almost 60% of all brain tumors in adults [123][81]. Gliosarcoma, a variant of GBM, is a highly aggressive malignant form of metastatic brain tumor. Although techniques based on tumor morphology (e.g., MRI, histopathology biopsy) to distinguish gliosarcoma from other GBMs, the early diagnosis of this aggressive disease is still poor, and the survival of patients after diagnosis is generally less than one year. Additionally, conventional treatment methods for GBM, such as radiotherapy, chemotherapy, and their combinations, are not effective. The abnormal activity of tumor cells and their resistance to chemotherapy and radiotherapy are major reasons for the higher fatality rate of GBM [124][82]. Thus, it would be a significant advance to discover specific molecules targeting gliosarcoma for early diagnosis and for treatments such the inhibition of GBM cell activity and increased radiosensitivity [125][83].
3. Conclusions and Future Perspective
Countries with rapidly aging populations will be challenged in the future by an increasing number of people affected by several neurodegenerative diseases. By 2050, over two billion people will be over 60 years old and the number of people over 80 will have tripled, from 137 million today to 425 million. This increase in the number of elderly individuals is expected to be accompanied by a proportional rise in the number of patients affected by neurological diseases. An increased incidence of brain tumors is also expected, both because cancer incidence increases with age and is possibly exacerbated by the diminished efficiency of repair mechanisms in the elderly brain. This rise in patients with neurological diseases and brain tumors could be diminished if factors that make the elderly susceptible to neurological disorders and the mechanisms of protein accumulation, impairment in degradation of aggregates, and neuronal cell death were understood, and appropriate diagnostics and treatments were developed. Therefore, understanding the fundamental bases of neurological diseases and brain tumors and the impact on these with aging will identify means of their prevention or cure and improve the quality of life for many people in old age. Antibodies are currently a primary means of diagnosis of molecular biomarkers of neurological disease, particularly for protein biomarkers. However, antibodies are expensive to produce, have batch-to-batch variation that requires extensive quality control, and they require refrigerated storage. When used therapeutically, antibodies also must first be “humanized” to avoid immune rejection. Opportunities for diagnosis and therapies can be expanded with the inclusion of aptamers that are specific for disease biomarkers and aptamer constructs that can be used to address therapeutic challenges. Although aptamers provide advantages and many new opportunities for diagnostic and therapeutic applications, their representation in modern diagnostics and therapeutics is low. Compared with antibodies, aptamers are relatively recently discovered molecules and their development into approved diagnostic and therapeutic agents will take time. In this review, we have summarized the available aptamers used for diagnosis and therapy and listed them with sequences and properties in Table 3. However, there are many more potential aptamer targets with links to neurological diseases that include LRRK2, Parkin, PINK1, DRP-1, DJ-1, UBQLN2, C9orf72, NEK-1, and FAS. With further optimization of their function and standardization of their characterization, the application of aptamers is expected to gain momentum and provide for many new opportunities in diagnostics and therapeutics in the field of neuroscience.Table 3.
Aptamers used in neuroscience applications.
| Aptamer | Sequence (5′-3′) | Target | Kd | Ref. | |
|---|---|---|---|---|---|
| BT5.6 | GGGGACGTAAATTGGATGTGGCTGCTTATGCTCTACTTG | BoNT-E | 53 nM | [47] | |
| M-30 | GGTATTGAGGGTCGCATCCCGTGGAAACAGGTTCATTGGGCGCAC TCCGCTTTCTGTAGATGGCTCTAACTCTCCTCT | saxitoxin | 128 nM | [73] | [84] |
| α-Tox-T2 | AGTTAGGGGCGACATGACCAAACGTT | α-toxin | 2.85 nM | [74] | [85] |
| Dopa2 | GCCGCGGAAGACGUUGGAAGGAUAGAUACCUACAACGGGGAAUAUAGAGGCCACCACAUAGUGAGGCCCUCCUCCCAAG | dopamine | 2.8 μM | [77] | [86] |
| T-SO508 | GCCTGTGGTGTTGGGGCGGGTGCG | amyloid beta | 68 nM | [91] | [87] |
| T-SO530 | GGTGCGGCGGGACTAGTGGGTGTG | amyloid beta | 63 nM | [91] | [87] |
| ssDNA1 | GCGGAGCGTGGCAGG | Tau381 | 190 nM | [94] | [88] |
| DNA aptamer | - | Tau441 | 28 nM | [96] | [89] |
| E2 | - | amyloid beta 1–40 | 10.9 μM | [97] | [90] |
| N2 | - | amyloid beta 1–40 | 21.6 μM | [97] | [90] |
| TH14 | CGCAACGCCGGGCCACTACGCGAATGGCAAGCCCGTCGAC | BACE1 | 280 nM | [101] | [91] |
| S10 | GTACACGTCGGCCACCTACGCGAAGTGGAAGCCTCATTTG | BACE1 | 360 nM | [101] | [91] |
| M5-15 | - | α-syn | - | [103] | [92] |
| AN58 | - | GluR2 | - | [116] | [74] |
| MS1 | AGGGGTGGGGAGGGGTGGGGA | huntingtin | - | [120] | [78] |
| MS2 | AGGGGTGGGGAGGGGAGGGGA | huntingtin | - | [120] | [78] |
| U2 | - | EGFRvIII | 6.27 nM | [125] | [83] |
| SLYC3 | CACTACAGAGGTTGCGTCTGTCCCACGTTGTCATGGGGGGTTGGCCTG | EpCAM | - | [142] | [93] |
| TEPN | GCGCGGTACCGCGCTAACGGATTCCTTTTCCGT | transferrin receptor | 65 nM | [142] | [93] |
References
- Rangel, A.E.; Chen, Z.; Ayele, T.M.; Heemstra, J.M. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 2018, 46, 8057–8068.
- Ilgu, M.; Nilsen-hamilton, M. Aptamers in Analytics. Analyst 2016, 141, 1551–1558.
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822.
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468.
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510.
- Blind, M.; Blank, M. Aptamer Selection Technology and Recent Advances. Mol. Ther. Nucleic Acids 2015, 4, e223.
- Kang, K.-N.N.; Lee, Y.-S.S. RNA aptamers: A review of recent trends and applications. Future Trends Biotechnol. 2013, 131, 153–169.
- Munzar, J.D.; Ng, A.; Juncker, D. Duplexed aptamers: History, design, theory, and application to biosensing. Chem. Soc. Rev. 2019, 48, 1390–1419.
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941.
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202.
- Boussebayle, A.; Groher, F.; Suess, B. RNA-based Capture-SELEX for the selection of small molecule-binding aptamers. Methods 2019, 161, 10–15.
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001, 294, 126–131.
- Oh, S.S.; Plakos, K.; Lou, X.; Xiao, Y.; Soh, H.T. In vitro selection of structure-switching, self-reporting aptamers. Proc. Natl. Acad. Sci. USA 2010, 107, 14053–14058.
- Golden, M.C.; Collins, B.D.; Willis, M.C.; Koch, T.H. Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. J. Biotechnol. 2000, 81, 167–178.
- Ohuchi, S. Cell-Selex technology. BioRes. Open Access 2012, 1, 265–272.
- Sola, M.; Menon, A.P.; Moreno, B.; Meraviglia-Crivelli, D.; Soldevilla, M.M.; Cartón-García, F.; Pastor, F. Aptamers Against Live Targets: Is In Vivo SELEX Finally Coming to the Edge? Mol. Ther. Nucleic Acids 2020, 21, 192–204.
- Wondergem, J.A.J.; Schiessel, H.; Tompitak, M. Performing SELEX experiments in silico. J. Chem. Phys. 2017, 147, 174101.
- Mosing, R.K.; Bowser, M.T. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX). Methods Mol. Biol. 2009, 535, 33–43.
- Vater, A.; Jarosch, F.; Buchner, K.; Klussmann, S. Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: Tailored-SELEX. Nucleic Acids Res. 2003, 31, e130.
- Lai, J.C.; Hong, C.Y. Magnetic-assisted rapid aptamer selection (MARAS) for generating high-affinity DNA aptamer using rotating magnetic fields. ACS Comb. Sci. 2014, 16, 321–327.
- Biondi, E.; Benner, S.A. Artificially expanded genetic information systems for new aptamer technologies. Biomedicines 2018, 6, 53.
- Breuers, S.; Bryant, L.L.; Legen, T.; Mayer, G. Robotic assisted generation of 2′-deoxy-2′-fluoro-modifed RNA aptamers—High performance enabling strategies in aptamer selection. Methods 2019, 161, 3–9.
- Szeto, K.; Latulippe, D.R.; Ozer, A.; Pagano, J.M.; White, B.S.; Shalloway, D.; Lis, J.T.; Craighead, H.G. RAPID-SELEX for RNA aptamers. PLoS ONE 2013, 8, e82667.
- Nguyen, V.T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple GO-SELEX for efficient screening of flexible aptamers. Chem. Commun. 2014, 50, 10513–10516.
- Ahn, J.Y.; Jo, M.; Dua, P.; Lee, D.K.; Kim, S. A sol-gel-based microfluidics system enhances the efficiency of RNA aptamer selection. Oligonucleotides 2011, 21, 93–100.
- Smith, J.D.; Gold, L. Conditional-Selex. U.S. Patent 650,688,7B1, 26 December 2002.
- Dausse, E.; Barré, A.; Aimé, A.; Groppi, A.; Rico, A.; Ainali, C.; Salgado, G.; Palau, W.; Daguerre, E.; Nikolski, M.; et al. Aptamer selection by direct microfluidic recovery and surface plasmon resonance evaluation. Biosens. Bioelectron. 2016, 80, 418–425.
- Burke, D.H.; Willis, J.H. Recombination, RNA evolution, and bifunctional RNA molecules isolated through chimeric SELEX. RNA 1998, 4, 1165–1175.
- Lecocq, S.; Spinella, K.; Dubois, B.; Lista, S.; Hampel, H.; Penner, G. Aptamers as biomarkers for neurological disorders. PLoS ONE 2018, 13, e0190212.
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434.
- Hoinka, J.; Zotenko, E.; Friedman, A.; Sauna, Z.E.; Przytycka, T.M. Identification of sequence-structure RNA binding motifs for SELEX-derived aptamers. Bioinformatics 2012, 28, i215–i223.
- Hoinka, J.; Berezhnoy, A.; Sauna, Z.E.; Gilboa, E.; Przytycka, T.M. AptaCluster—A method to cluster HT-SELEX aptamer pools and lessons from its application. In International Conference on Research in Computational Molecular Biology; Springer: Cham, Switzerland, 2014.
- Ortigao, J.F.R.; Rösch, H.; Montenarh, M.; Fröhlich, A.; Seliger, H. Oligonucleotide Analogs with Terminal 3′,3′- and 5′,5′-Internucleotidic Linkages as Antisense Inhibitors of Viral Replication. Antisense Res. Dev. 1991, 1, 380.
- Dougan, H.; Lyster, D.M.; Vo, C.V.; Stafford, A.; Weitz, J.I.; Hobbs, J.B. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl. Med. Biol. 2000, 27, 289–297.
- Padilla, R.; Sousa, R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutant T7 RNA polymerase (RNAP). Nucleic Acids Res. 1999, 27, 1561–1563.
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjić, N. 2′-fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165): Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567.
- Saccà, B.; Lacroix, L.; Mergny, J.L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005, 33, 1182–1192.
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123, 1032–1039.
- Pozmogova, G.E.; Zaitseva, M.A.; Smirnov, I.P.; Shvachko, A.G.; Murina, M.A.; Sergeenko, V.I. Anticoagulant effects of thioanalogs of thrombin-binding DNA-aptamer and their stability in the plasma. Bull. Exp. Biol. Med. 2010, 150, 180–184.
- Lee, C.H.; Lee, S.H.; Kim, J.H.; Noh, Y.H.; Noh, G.J.; Lee, S.W. Pharmacokinetics of a Cholesterol-conjugated Aptamer Against the Hepatitis C Virus (HCV) NS5B Protein. Mol. Ther. Nucleic Acids 2015, 4, e254.
- Willis, M.C.; Collins, B.; Zhang, T.; Green, L.S.; Sebesta, D.P.; Bell, C.; Kellogg, E.; Gill, S.C.; Magallanez, A.; Knauer, S.; et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug. Chem. 1998, 9, 573–582.
- Da Pieve, C.; Blackshaw, E.; Missailidis, S.; Perkins, A.C. PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice. Bioconjug. Chem. 2012, 23, 1377–1381.
- Trinh, T.; Zhu, G.; Xiao, X.; Puszyk, W.; Sefah, K.; Wu, Q.; Tan, W.; Liu, C. A synthetic aptamer-drug adduct for targeted liver cancer therapy. PLoS ONE 2015, 10, e0136673.
- Kato, K.; Ikeda, H.; Miyakawa, S.; Futakawa, S.; Nonaka, Y.; Fujiwara, M.; Okudaira, S.; Kano, K.; Aoki, J.; Morita, J.; et al. Structural basis for specific inhibition of Autotaxin by a DNA aptamer. Nat. Struct. Mol. Biol. 2016, 23, 395–401.
- Lim, Y.C.; Kouzani, A.Z.; Duan, W. Aptasensors: A review. J. Biomed. Nanotechnol. 2010, 6, 93–105.
- Hu, M.; Zhang, K. The application of aptamers in cancer research: An up-to-date review. Future Oncol. 2013, 9, 369–376.
- Ren, S.; Shin, H.S.; Gedi, V.; Dua, P.; Lee, D.K.; Kim, S. Selection of DNA Aptamers Against Botulinum Neurotoxin E for Development of Fluorescent Aptasensor. Bull. Korean Chem. Soc. 2017, 38, 324–328.
- Kim, T.H.; Lee, S.W. Aptamers for anti-viral therapeutics and diagnostics. Int. J. Mol. Sci. 2021, 22, 4168.
- Marrazza, G. Aptamer Sensors. Biosensors 2017, 7, 5.
- Buglak, A.A.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. Methods and applications of in silico aptamer design and modeling. Int. J. Mol. Sci. 2020, 21, 8420.
- Morita, Y.; Leslie, M.; Kameyama, H.; Volk, D.E.; Tanaka, T. Aptamer therapeutics in cancer: Current and future. Cancers 2018, 10, 80.
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117.
- Davis, K.A.; Abrams, B.; Lin, Y.; Jayasena, S.D. Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res. 1996, 24, 702–706.
- Kaur, H.; Shorie, M. Nanomaterial based aptasensors for clinical and environmental diagnostic applications. Nanoscale Adv. 2019, 1, 2123–2138.
- Kou, X.; Zhang, X.; Shao, X.; Jiang, C.; Ning, L. Recent advances in optical aptasensor technology for amplification strategies in cancer diagnostics. Anal. Bioanal. Chem. 2020, 412, 6691–6705.
- Yan, S.R.; Foroughi, M.M.; Safaei, M.; Jahani, S.; Ebrahimpour, N.; Borhani, F.; Rezaei Zade Baravati, N.; Aramesh-Boroujeni, Z.; Foong, L.K. A review: Recent advances in ultrasensitive and highly specific recognition aptasensors with various detection strategies. Int. J. Biol. Macromol. 2020, 155, 184–207.
- Banerjee, J.; Nilsen-Hamilton, M. Aptamers: Multifunctional molecules for biomedical research. J. Mol. Med. 2013, 91, 1333–1342.
- Catuogno, S.; Esposito, C.L. Aptamer cell-based selection: Overview and advances. Biomedicines 2017, 5, 49.
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the therapeutics and diagnostics pipelines. Theranostics 2018, 8, 4016–4032.
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. N. Drugs 2014, 32, 178–187.
- Catuogno, S.; Esposito, C.L.; de Franciscis, V. Aptamer-mediated targeted delivery of therapeutics: An update. Pharmaceuticals 2016, 9, 69.
- Chandola, C.; Neerathilingam, M. Aptamers for Targeted Delivery: Current Challenges and Future Opportunities. In Role of Novel Drug Delivery Vehicles in Nanobiomedicine; BoD–Books on Demand: Norderstedt, Germany, 2020.
- Bayrac, A.T.; Sefah, K.; Parekh, P.; Bayrac, C.; Gulbakan, B.; Oktem, H.A.; Tan, W. In vitro selection of DNA aptamers to glioblastoma multiforme. ACS Chem. Neurosci. 2011, 2, 175–181.
- de Almeida, C.E.B.; Alves, L.N.; Rocha, H.F.; Cabral-Neto, J.B.; Missailidis, S. Aptamer delivery of siRNA, radiopharmaceutics and chemotherapy agents in cancer. Int. J. Pharm. 2017, 525, 334–342.
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015.
- Sivakumar, P.; Kim, S.; Kang, H.C.; Shim, M.S. Targeted siRNA delivery using aptamer-siRNA chimeras and aptamer-conjugated nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, 1–20.
- WHO. Dementia. 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 30 September 2021).
- Bouvier-Müller, A.; Ducongé, F. Nucleic acid aptamers for neurodegenerative diseases. Biochimie 2018, 145, 73–83.
- Qu, J.; Yu, S.; Zheng, Y.; Zheng, Y.; Yang, H.; Zhang, J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2017, 74, 683–695.
- Lange, C.W.; VanBrocklin, H.F.; Taylor, S.E. Photoconjugation of 3-azido-5-nitrobenzyl-fluoride to an oligonucleotide aptamer. J. Label. Compd. Radiopharm. 2002, 45, 257–268.
- Pollak, T.A.; Rogers, J.P.; Nagele, R.G.; Peakman, M.; Stone, J.M.; David, A.S.; McGuire, P. Antibodies in the diagnosis, prognosis, and prediction of psychotic disorders. Schizophr. Bull. 2019, 45, 233–246.
- Mroczko, B.; Groblewska, M.; Litman-Zawadzka, A. The role of protein misfolding and tau oligomers (TauOs) in Alzheimer’s disease (AD). Int. J. Mol. Sci. 2019, 20, 4661.
- Wallin, M.T.; Culpepper, W.J.; Nichols, E.; Bhutta, Z.A.; Gebrehiwot, T.T.; Hay, S.I.; Khalil, I.A.; Krohn, K.J.; Liang, X.; Naghavi, M.; et al. Global, regional, and national burden of multiple sclerosis 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 269–285.
- Huang, Z.; Pei, W.; Jayaseelan, S.; Shi, H.; Niu, L. RNA Aptamers Selected against the GluR2 Glutamate Receptor Channel. Biochemistry 2007, 46, 12648–12655.
- Zacco, E.; Graña-Montes, R.; Martin, S.R.; de Groot, N.S.; Alfano, C.; Tartaglia, G.G.; Pastore, A. RNA as a key factor in driving or preventing self-assembly of the TAR DNA-binding protein 43. J. Mol. Biol. 2019, 431, 1671–1688.
- Huang, W.J.; Chen, W.W.; Zhang, X. Huntington’s disease: Molecular basis of pathology and status of current therapeutic approaches. Exp. Ther. Med. 2016, 12, 1951–1956.
- Chaudhary, R.K.; Patel, K.A.; Patel, M.K.; Joshi, R.H.; Roy, I. Inhibition of Aggregation of Mutant Huntingtin by Nucleic Acid Aptamers in Vitro and in a Yeast Model of Huntington’s Disease. Mol. Ther. 2015, 23, 1912–1926.
- Shin, B.; Jung, R.; Oh, H.; Owens, G.E.; Lee, H.; Kwak, S.; Lee, R.; Cotman, S.L.; Lee, J.M.; MacDonald, M.E.; et al. Novel DNA Aptamers that Bind to Mutant Huntingtin and Modify Its Activity. Mol. Ther. Nucleic Acids 2018, 11, 416–428.
- Macedo, B.; Cordeiro, Y. Unraveling prion protein interactions with aptamers and other PrP-binding nucleic acids. Int. J. Mol. Sci. 2017, 18, 1023.
- Cavaliere, P.; Pagano, B.; Granata, V.; Prigent, S.; Rezaei, H.; Giancola, C.; Zagari, A. Cross-talk between prion protein and quadruplex-forming nucleic acids: A dynamic complex formation. Nucleic Acids Res. 2013, 41, 327–339.
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9.
- Bastien, J.I.L.; McNeill, K.A.; Fine, H.A. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer 2015, 121, 502–516.
- Zhang, X.; Peng, L.; Liang, Z.; Kou, Z.; Chen, Y.; Shi, G.; Li, X.; Liang, Y.; Wang, F.; Shi, Y. Effects of Aptamer to U87-EGFRvIII Cells on the Proliferation, Radiosensitivity, and Radiotherapy of Glioblastoma Cells. Mol. Ther. Nucleic Acids 2018, 10, 438–449.
- Gao, S.; Zheng, X.; Wu, J. A biolayer interferometry-based competitive biosensor for rapid and sensitive detection of saxitoxin. Sens. Actuators B Chem. 2017, 246, 169–174.
- Dhiman, A.; Anand, A.; Malhotra, A.; Khan, E.; Santra, V.; Kumar, A.; Sharma, T.K. Rational truncation of aptamer for cross-species application to detect krait envenomation. Sci. Rep. 2018, 8, 17795.
- Mannironi, C.; Di Nardo, A.; Fruscoloni, P.; Tocchini-Valentini, G.P. In vitro selection of dopamine RNA ligands. Biochemistry 1997, 36, 9726–9734.
- Tsukakoshi, K.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers that recognize alpha-synuclein oligomers using a competitive screening method. Anal. Chem. 2012, 84, 5542–5547.
- Krylova, S.M.; Musheev, M.; Nutiu, R.; Li, Y.; Lee, G.; Krylov, S.N. Tau protein binds single-stranded DNA sequence specifically—The proof obtained in vitro with non-equilibrium capillary electrophoresis of equilibrium mixtures. FEBS Lett. 2005, 579, 1371–1375.
- Lisi, S.; Fiore, E.; Scarano, S.; Pascale, E.; Boehman, Y.; Ducongé, F.; Chierici, S.; Minunni, M.; Peyrin, E.; Ravelet, C. Non-SELEX isolation of DNA aptamers for the homogeneous-phase fluorescence anisotropy sensing of tau Proteins. Anal. Chim. Acta 2018, 1038, 173–181.
- Takahashi, T.; Tada, K.; Mihara, H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol. Biosyst. 2009, 5, 986–991.
- Rentmeister, A.; Bill, A.; Wahle, T.; Walter, J.; Famulok, M. RNA aptamers selectively modulate protein recruitment to the cytoplasmic domain of b -secretase BACE1 in vitro. RNA 2006, 1650–1660.
- Tsukakoshi, K.; Harada, R.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer which binds to α-synuclein. Biotechnol. Lett. 2010, 32, 643–648.
- Macdonald, J.; Henri, J.; Goodman, L.; Xiang, D.; Duan, W.; Shigdar, S. Development of a Bifunctional Aptamer Targeting the Transferrin Receptor and Epithelial Cell Adhesion Molecule (EpCAM) for the Treatment of Brain Cancer Metastases. ACS Chem. Neurosci. 2017, 8, 777–784.
More
