Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Peter Tang and Version 1 by Sandra N. Pinto.
Bacterial biofilms are defined as complex aggregates of bacteria that grow attached to surfaces or are associated with interfaces. Bacteria within biofilms are embedded in a self-produced extracellular matrix made of polysaccharides, nucleic acids, and proteins. It is recognized that bacterial biofilms are responsible for the majority of microbial infections that occur in the human body, and that biofilm-related infections are extremely difficult to treat. This is related with the fact that microbial cells in biofilms exhibit increased resistance levels to antibiotics in comparison with planktonic (free-floating) cells.
- bacterial biofilms
- biofilm detection
- biofilm imaging
- antimicrobial treatment
- antibiofilm agents
1. Introduction
Antimicrobial resistance is the inevitable consequence of prescribing antibiotics, and bacteria will continue to develop resistance to therapies. This is a critical problem in hospital environments in particular due to the low numbers of novel compounds or strategies under development [1]. Several factors contribute to this scarcity, including a market failure, produced by the lack of incentive for pharmaceutical companies to develop antibiotics. This lack of incentives for development of novel antibiotics is a result of a low return on investment, since they are fast-acting drugs, use of novel antibiotics is often reserved, and their use is ultimately unpredictable as resistance evolves [1].
A 2014 report commissioned by the U.K. government also predicted that millions of people are expected to die prematurely because of drug resistance over the next 35 years worldwide and the world’s gross domestic product (GDP) will be 2 to 3.5% lower than it otherwise would be in 2050 [2].
This problem is even more relevant when is associated with bacterial biofilms, since bacteria growing in biofilms show significantly reduced antibiotic susceptibility. For instance, implant-related infections are very hard to treat [3]. This is due to the fact that clinical procedures to treat implant-related infections involve replacement of the implant that can increase the risk for the patient developing severe complications, as well as the very high costs associated with this procedure [4].
Bacterial biofilms consist of densely packed communities that grow attached to surfaces and are responsible for the majority of human clinical infections [5,6][5][6]. Their resistance to conventional antibiotics is 10- to 1000-fold higher than that of planktonic bacteria [7].
This increased resistance can be attributed to different factors, including decreased diffusion of antimicrobial agents through the self-produced extracellular matrix (made of polysaccharides, nucleic acids, and proteins matrix) [8[8][9][10],9,10], altered metabolic activity, and formation of persister cells [11]. Moreover, a relevant characteristic of biofilms is the fact that bacteria are able to inter-communicate and collaborate to survive in even the hardest conditions. This cell–cell communication mechanism is known as the quorum sensing (QS) system [12]. Bacteria QS system consists of self-produced extracellular chemical signals (called autoinducers), and it can control important virulence factors such as biofilm formation and maturation, antibiotic resistance, bacterial swarming, and bacteria–host interaction [12,13][12][13].
2. Imaging of Biofilms and the Diversity of Detection Methods
Currently, nuclear medicine imaging is still the standard technique for the detection of infectious diseases [14]. However, this technique has several drawbacks, including the fact that (i) it involves the exposition of the patient to radiation, (ii) it requires specialized equipment, and (iii) it requires operator training.
As recognized, there is an urgency to develop more accurate diagnostic tools and treatment, particularly when bacterial infection progresses to biofilm. The noninvasive technique in clinical use does not offer an optimized approach to detect biofilm infection. The low resolution, low practicality, and impossibility to distinguish between bacterial infection and sterile inflammation are the reasons to develop new diagnostics tools for biofilm detection in clinical environments [15]. In the last few years, development of diagnostic approaches became urgent in order to improve the treatment of bacterial infections and preserve some medical procedures that need alternative tools to prevent and treat bacterial infections.
2.1. Nuclear Imaging
Nuclear imaging has been applied in oncology and in infectious disease diagnostics. Some radionuclides such as technetium-99m, iodide-125, and indium-111 have been shown for years to be useful tools to radiolabeled compounds for medical applications [16].
However, some disadvantages are found in common bacterial imaging agents such as difficult radiochemical synthesis, non-specific adsorption, or small target receptor expression on bacteria of interest [16]. Despite these limitations, recent works have revealed that there are bacterial metabolites that can be radiolabeled and used as tracers to identify bacterial biofilm infections, such as the maltodextrin transport system [17,18][17][18].
The carbohydrate transport and metabolism has been reported as an essential tool for proliferation of bacteria in the human organism. Thus, there are some strategies that include carbohydrate metabolism as a target that can be very helpful in improving nuclear imaging in the field of infectious disease diagnostics. For instance, 18FDG (fluorodeoxyglucose) is one example of an important radiopharmaceutical that has been used for many years on positron emission tomography (PET). However, 18FDG as a contrast agent shows a high uptake in mammalian cells and absence of distinguishing bacterial infections from cancer or inflammation [19,20][19][20].
Although efforts are being made to find new radiopharmaceutical and contrast agents, there are some complications associated with radiochemical synthesis and low affinity/specify at the bacteria target. With the aim of increasing the sensitivity of currently imaging methods, researchers have developed other contrast agents targeting the bacterial carbohydrate metabolism. This is the case of 18F-maltohexaose (MH18F) [17]. MH18F nuclear imaging agent is internalized by a bacteria-specific maltodextrin transporter. Thus, the contrast agents conjugated with maltohexose were only internalized by bacterial cells and not by mammalian cells, which do not express maltodextrin transporters [21]. Moreover, one of the major advantages of maltodextrin-based compounds is their lower toxicity because they are widely used as food additives. The development of nuclear agents such as MH18F might be crucial to bacterial biofilm detection at an early stage.
2.2. Ultrasound Contrast Agent Imaging
Ultrasonic imaging techniques and their combination with other methods have been explored in order to enhance the strategies to detect and quantify early and mature biofilms [22,23][22][23]. The acoustic approach has the advantage of monitoring the surface biofouling in real time, and it has been proved that the ultrasonic technique can monitor formation and growth of some microbiological colonies [22].
Ultrasound medical imaging has been developed with the addition of contrast agents, especially encapsulated gas bubbles, which has led to an improvement in medical diagnosis [24]. Another improvement was a novel design of ultrasound contrast agents (UCAs), including a target ligand to establish the difference between infectious and healthy tissue [25].
It is important to have a detection technique that allows for identification of biofilms in early stages because the late diagnostic of mature biofilms sometimes compromises their clinical treatment inside the human body. Echocardiography, for instance, has several limitations in the detection of intra-cardiac biofilm. Staphylococcus aureus is the most common isolate in infective endocarditis, and strategies have been developed that can evaluate and characterize mechanical and structural properties of its biofilm. In this sense, researchers have developed strategies that could be evaluated and that characterize mechanical and structural properties of S. aureus biofilm [26]; this is because S. aureus is the most common isolate in infective endocarditis [27]. S. aureus biofilm has been studied through a combination of targeted ultrasound contrast agents (UCAs) and fluorescent probes. In the first step, developed UCAs that bind a carbohydrate epitope allowed for a spatial scanning of biofilm structure by high-frequency scanning acoustic microscopy (SAM). A complementary analysis occurred with TRITC-labelled streptavidin by fluorescence microscopy. The merge between high-frequency acoustic scanning and fluorescence imaging allowed for the acquisition of spatial resolution and detection of the biofilm components [26]. The results obtained showed an improvement in biofilm diagnostics.
2.3. Optical Imaging and Probes
Optical imaging offers an important tool to understand/visualize the 3D biofilm structure. Multiple techniques are included in this range, such as scanning electron microscopy (SEM), confocal scanning laser microscopy (CSLM), light microscopy, infrared spectroscopy, and reflectance spectroscopy [28]. Figure 1 illustrates a S. aureus biofilm visualized with the use of CSLM and SEM.
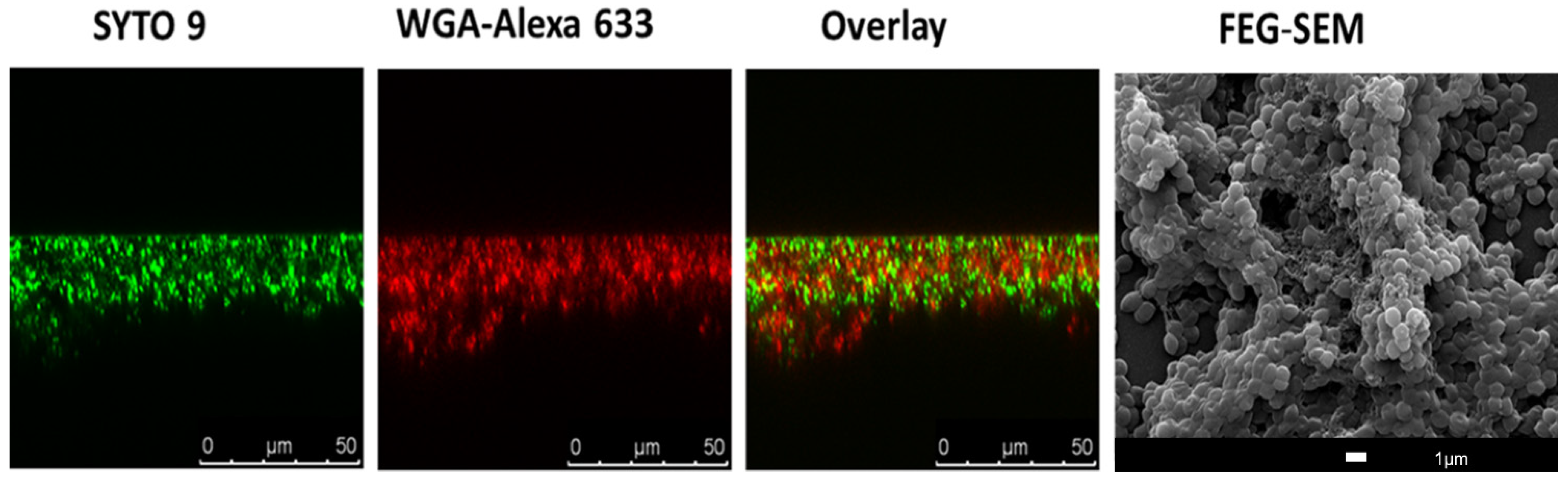
Figure 1. Imaging of bacterial biofilms with confocal scanning laser microscopy (CSLM) and SEM. Twenty-four hour S. aureus JE2 (MRSA) biofilm was co-labelled with SYTO 9 (green channel), a nucleic acid binding dye, and with WGA-ALEXA 633 (red channel), a wheat germ agglutinin dye that labels S. aureus biofilm matrix. The overlay between the two channels is also represented. In the right image, there is a representation scanning electron image (SEM) of 24 h S. aureus JE2 biofilms.
SEM, a technique with high resolution that is based on surface scattering and absorption of electrons, has been used in different studies to visualize biofilms (Figure 1) since it is able to detect key structural components such as the presence of biofilm matrix [29,30][29][30]. Moreover, researchers have been using SEM to evaluate the efficacy of anti-biofilm compounds [31,32,33][31][32][33]. However, SEM is a very expensive technique, and quantitation of the biofilm is rather difficult, including the fact that researchers cannot work with live samples.
Due to this, the most common used methodology to study the 3D biofilm morphology of biofilms is probably CSLM, and in fact, it is recognized that CSLM represents an important advance in technology-associated biofilm imaging [34]. In CSLM, due to the presence of a confocal pinhole, the out-of-focus fluorescent signals are eliminated [35], which is relevant when it is considered for instance with traditional fluorescent microscopy. Moreover, it allows for the formation of high-resolution images at different depths [36], which is crucial in biofilm studies. The tridimensional morphology and physiology of biofilms can then be screened by CLSM using a combination of molecular probes and fluorescent proteins optimized to target/visualize biofilm components. Most probes and fluorescent proteins are designed to stain cellular organelles and structures. However, in the last decades, there has been an effort in the development of proteins and fluorochromes to target, for instance, the extracellular matrix of biofilms. This includes the application of fluorescently labelled lectins (Figure 1) to visualize and characterize the biofilm matrix, in particular the extracellular polysaccharide components [37,38,39][37][38][39].
Extracellular DNA (eDNA) is also often a target for extracellular matrix imaging using CSLM. Propidium iodide, TOTO-1, and TO-PRO-3 iodide are probes that were used in this context, providing excellent distinction between biofilm eDNA component and the intracellular DNA found in biofilm cells [8,40][8][40]. These probes are often used in combination with SYTO 9 [8] or SYTO 60. SYTO 9 is a green fluorescent nucleic acid-binding dye. The fact that SYTO 9 is a membrane-permeable probe and TO-PRO-3 iodide or propidium iodide can only label cells with damaged membranes allows the viewer to discriminate between viable and nonviable cells [8].
Other fluorescent probes for extracellular DNA (eDNA) detection have recently been developed. This is the case of CDr15 probe, which was evaluated on Pseudomonas aeruginosa ΔwspF with a highly elevated cyclic-di-GMP content (mimicking the biofilm mode of growth) and a pYhjH strain with a low intracellular cyclic-di- GMP content (representative of the planktonic mode of growth). The results showed that CDr15 probes bind effectively to eDNA. The robustness of CDr15 as a diagnostic in vivo probe was evaluated on corneal infection model, and the results showed that biofilm regions were visualized after CDr15 treatment [41].
Identification of novel fluorescent probes together with targeting different biofilm structures will greatly facilitate diagnosis of biofilm infection. In this sense, a fluorescent probe, CDy11, that targets amyloid-like fibers in the P. aeruginosa biofilm matrix was developed. It was demonstrated that CDy11 allows for detection using in vivo imaging of P. aeruginosa in implant and corneal infection mice models [42]. In addition, CDy14 was identified as a potential fluorescent probe to target Psl exopolysaccharide in P. aeruginosa [43]. In this context, amphiphilic fluorescent carbon dots were developed and applied to assist the characterization of bacterial biofilm matrix [44]. The amphiphilic carbon dots (C-dots) were shown to readily bind to the EPS scaffold of P. aeruginosa, and it was detected for the first time as a dendritic morphology of the EPS.
Furthermore, the peptide nucleic acid fluorescence in situ hybridization (PNA FISH) technique has also been used to study biofilm’s structure and composition. Traditional FISH is a molecular technique on which labeled DNA probes hybridize to their complementary nucleic acid targets. The use of FISH (namely, DNA probes) to study microorganisms and biofilms can lead to some drawbacks, including poor target site specificity and poor signal-to-noise ratio [45,46][45][46]. The limitations associated with FISH can be overcome with the use of peptide nucleic acid (PNA) probes; PNA is a synthetic DNA analogue with a stronger binding to nucleic acids [47]. PNA FISH technique is very helpful for the CLSM observation of mixed biofilms since it allows for the use of multiple fluorescent probe labels that are characteristic of a specific microorganism [48,49][48][49].
In vivo biofilm detection possesses a challenge for the scientific community. One promising approach for this purpose relies on the use of laser capture microdissection (LCM). Laser capture microdissection is a high-resolution technique that allows researchers to rapidly sample/isolate individual cells or cell compartments from solid tissue with the aid of a laser beam [62,63][50][51]. LCM has also been used to isolate non-cellular structures including amyloid plaques [64][52]. This microdissection technique is often used in cancer research, e.g., [65][53], and now researchers are using it to obtain information related with in vivo biofilms. For instance, very recently, the adaptation of B. cereus in G. mellonella gut infection model was demonstrated for the first time with LCM [66][54].
Another valuable imaging technique for identification of in vivo biofilms is target fluorescent imaging (TFLI). The principle of TFLI is targeting fluorophores that emit light outside the absorbance window of tissue in the near infrared region. There are some reports of targeting fluorescent imaging for tumor diagnostics, and the first clinical TFLI approach employment was observed in ovarian cancer surgery [67][55]. Furthermore, some studies have been published to also demonstrate the ability of TFLI for in vivo detection of bacteria [68,69][56][57]. Since TFLI emerged as useful tool for multiple diagnosis in clinical research, Marleen van Osteen and colleagues decided to combine the TFLI advantages with vancomycin’s well-known biodistribution profile. In this sense, the authors developed vanc-800CW as a new conjugate for optical biofilm imaging. For this propose the authors conjugated vancomycin with IRDye-800CW, a near-infrared fluorophore. The images were obtained by IVIS Lumina II imaging system [70][58].
The vanc-800CW potential as a fluorescent probe was evaluated in multiple models. The in vitro studies performed demonstrate a good detection for Streptococcus and Dermabacter species and minor detection of Corynebacterium. As expected, the results also confirmed the lack of vancomycin staining for Gram-negative bacteria such as P. aeruginosa and Escherichia coli [70][58].
To understand the potential of vanc-800CW in vivo model, the authors selected a mouse model of myositis induced by bioluminescent S. aureus. The administration of vanc-800CW allows for distinguishing between S. aureus-induced infection from E. coli induced-infection and sterile inflammation. The biodistribution profile also shows similarities with what is described for “native” vancomycin. A complementary post-mortem with contaminated implants was also performed to ensure the feasibility of BAI detection. The results were promising and confirmed the ability of vanc-88CW to stain Staphylococcus epidermis-containing implants. The fluorescent conjugated developed by Marleen van Osteen and colleagues displayed important and crucial results in biofilm imaging [70][58].
The application of carbon nanotube probes is another promising tool for in vivo targeting and fluorescence optical imaging of bacterial infections [71][59]. Using genetically engineered M13 virus as a multifunctional vector, Bardhan et al. synthesized NIR-II fluorescent SWNT probes, with additional functionalization on the virus for active targeting of bacterial infections [71][59]. The authors were able to successfully preform the detection of deep-tissue infective endocarditis using the SWNT probe.
2.4. Biofilm Detection with iTRAQ (Isobaric Tags for Relative and Absolute Quantitation)-Based Quantitative Proteomics Methods
As explained before, the biofilm structure contains several proteins that are important for its stability and maintenance [6]. The proteins present in the biofilm naturally depend not only on the type of pathogen but also on the developmental stage of the biofilm [72][60]. Therefore, identifying biofilm proteins can be a very useful biofilm detection method. For this purpose, isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative proteomics technique has been reported in several studies [73][61]. The iTRAQ technique allows for the identification and quantification of hundreds of proteins in different biological samples in one single experiment. It consists of the relative quantification with mass spectrometry of proteins in complex mixtures. iTRAQ technology uses isobaric reagents to label the primary amines of peptides and proteins [73][61]. During the iTRAQ process, reagents are reactive with amine groups, marking the sample peptides and maintaining the isobaric balance (sample mass does not change) [73,74][61][62]. An analysis of the reporter groups that are generated upon fragmentation in the mass spectrometer is then carried out. This procedure is commonly used to distinguish between normal and “diseased” samples and was also used to identify bacterial biofilm proteins [75,76][63][64]. Recently, an iTRAQ-based quantitative proteomics approach was used to identify protein markers associated with the biofilm formation of Enterococcus faecalis [77][65]. In this case, it was observed by iTRAQ that strong biofilm-forming clinical isolates have proteins associated with shikimate kinase pathway and sulfate transport upregulated. This is a relevant information since it can lead to the development of therapies that can act on these metabolic pathways, and consequently inhibit the biofilm formation of Enterococcus faecalis [77][65]. The iTRAQ technique has also been used to identify proteins present in biofilms that promote caries and other dental problems [78,79][66][67]. iTRAQ reporters determined that biofilm cells of Tannerella forsythia have upregulated oxidative stress response proteins, which is related with the fact that this sub-gingival pathogen is more resistant to oxidative stress, thus allowing it to persist in the oral cavity [79][67]. Thus, the iTRAQ-based quantitative proteomics technique can be very useful for biofilm detection and to find possible targets that could lead to biofilm eradication, as it allows for the understanding of which proteins and metabolic pathways are important for biofilm formation.
2.5. The Use of Artificial Intelligence (AI) Technology for Biofilm Detection
Machine learning, together with image processing, has been employed in recent years to assist doctors during clinical and diagnostic process [80,81][68][69].
For biofilm detection, the use of machine learning models was already reported, e.g., detection of E. coli biofilm using an electro-chemical impedance spectroscopy (EIS)-based biosensor [82][70]. Machine learning systems, for instance, can be trained to recognize multiple impedimetric parameters and determine bacteria concentration. The conjugation of machine learning systems with EIS already showed promising results, even with thicker biofilm [82][70].
Convolutional neural network (CNN) has already been reported as a successful deep learning model for improving diagnostic field [83][71]. The CNN model is trained to learn visual patterns from images and has been used for medical images recognition [84,85][72][73]. Recently, this model was tested to improve a rhinocytology diagnostic exam [81,86][69][74]. For instance, it allowed for the detection of the presence of biofilm on rhino-cytological scans. The sample was stained, and cyan-colored spots were observed and were directly related with biofilm infection [86][74]. The cyan spots can vary with stage/maturity of the biofilm, and the CNN model system can be trained to recognize these patterns. The CNN model was also applied for detection of biofilm formation (all four stages) attached onto a metallic material. To achieve this purpose, the researchers trained the system to recognize the main features of the process on the basis of microscopy features. For E. coli strain, this mathematical model showed results in accordance with experimental detection of metal biofilm [87][75].
Moreover, the CNN deep learning model can also be trained to detect polymicrobial biofilm. Antoine Buetti-Dinh et al. reported a CNN model trained to detect a biofilm composed by A. caldus strain, L. ferriphilum strain, and S. thermosulfi-dooxidans of sulfide minerals. When compared to human experts, the CNN model showed a 90% of accuracy in contrast with 50%, thus offering an accurate alternative to classical and time-consuming biochemical methods [83][71].
3. Antibacterial and Antibiofilm Strategies
The difficulty of treating bacterial biofilm-associated infections, as explained above, is highly associated with the recalcitrant character of bacterial biofilm and the development of resistance toward antibiotics. Therefore, the development of strategies that allow for efficient inhibition of biofilm formation and/or to completely eradicate biofilms is one of the most challenging research topics of the present day. Understanding the mechanism behind biofilm formation is crucial to developing potential control strategies. This includes exploring potential targets against c-di-GMP, extracellular polysaccharide and eDNA present in biofilm matrix, and bacterial cell membrane and biofilm quorum sensing. In addition, ribonucleases and small non-coding RNAs (sRNAs) can also in the future be considered as potential targets since several research papers were able to demonstrate their importance in biofilm formation and regulation (e.g., [88][76]). In fact, ribonucleases were found to affect biofilm formation in several bacteria, such as E. coli, Salmonella Typhimurium, P. aeruginosa, and Bacillus subtilis [88,89,90,91][76][77][78][79]. These RNA-degrading enzymes affect biofilms by controlling the expression of biofilm matrix genes but also by modulating the levels of c-di-GMP and other biofilm regulators [92,93,94][80][81][82]. Other RNA regulators, namely, sRNAs, have been found to have a very important role in biofilm formation and antibiotic resistance [95,96][83][84]. However, thus far, no therapeutic drugs target these biofilm regulators, mainly due to the lack of basic knowledge on how exactly ribonucleases and sRNAs could be used to disrupt biofilms.
In the context of therapeutic drugs, several compounds are currently being screened, including the antibody MEDI4893 and the antimicrobial peptide POL7080, both in clinical trial phase 2 [97][85]. Furthermore, in silico analysis or machine learning methods are becoming attractive strategies to help identify potential antibacterial and anti-biofilm inhibitory molecules [98,99,100][86][87][88]. In recent years, computational methods have emerged following the need for less consuming and more accurate results for the identification of antimicrobial and anti-biofilm molecules. In this context, some computational databases such as biofilm-active AMPs (BaAMPs) and tools/platforms including aBiofilm and Molib were created [101,102,103,104][89][90][91][92]. The aBiofilm platform has already provided the prediction of antimicrobial chemical molecules and their inhibitory activity [103][91]. Meanwhile, Molib tool, which is a training dataset of biofilm inhibitory molecules, has been shown to be even more accurate than the aBiofilm tool [104][92]. This focus on discovering small molecules with artificial intelligence might offer future solutions for the search of effective anti-biofilm drug discovery (see also Figure 2).
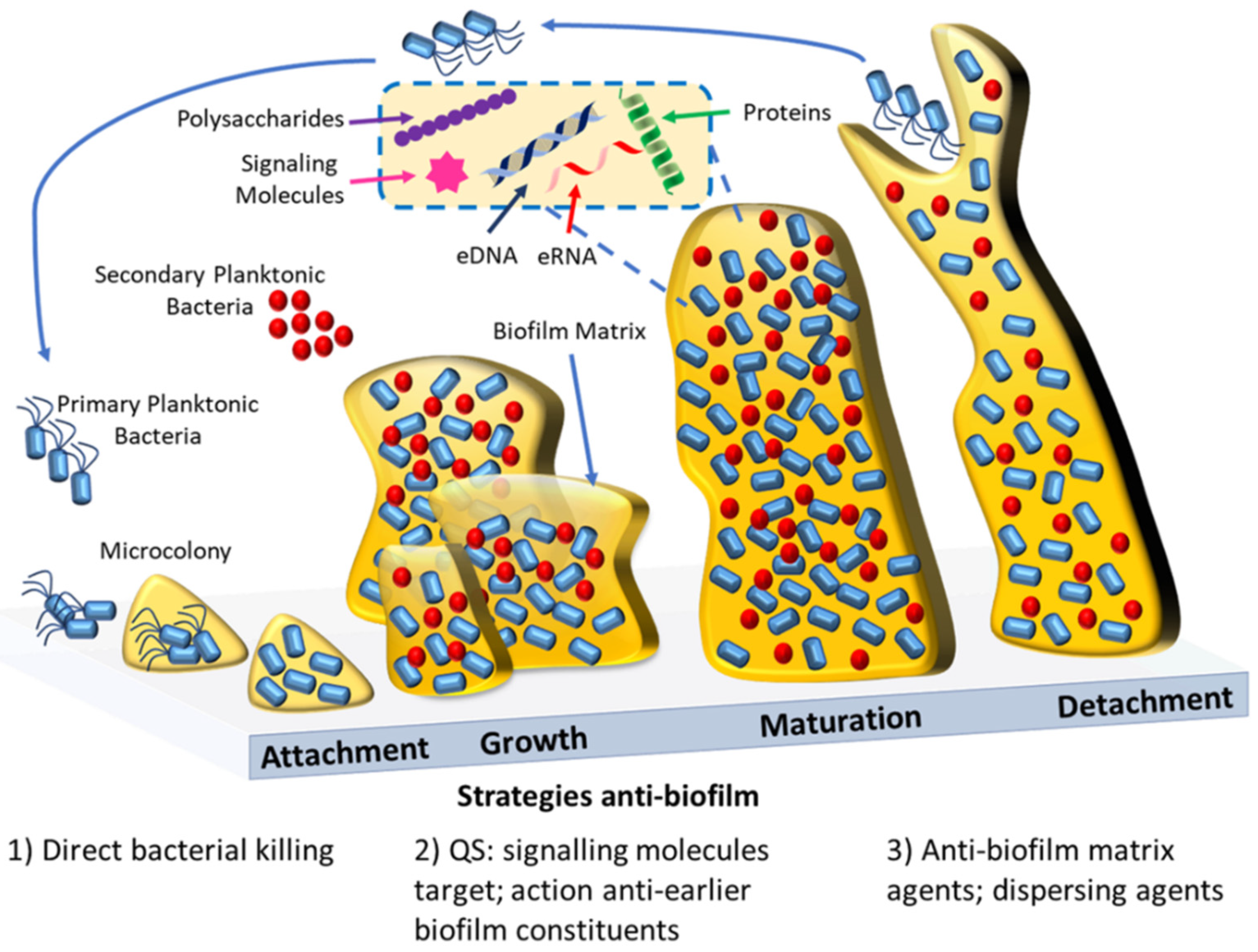
Figure 2. Schematic representation of biofilm formation and the current antibiofilm strategies. Biofilm formation starts with the initial reversible attachment of bacterial cells to a surface, then follows the growth of the biofilm within a matrix; maturation of the biofilm; and finally, when the environment conditions cease to be ideal, the reversal of the attachment with the dispersion of the cells that will colonize other superficies. Antibiofilm agents are capable of inhibiting the biofilm formation by bacteriostatic effects (1), e.g., antimicrobial polymers, or by acting against important early biofilm constituents (2), e.g., quorum sensing inhibitors. Mature biofilm can be disrupted by direct action against the biofilm matrix (3), e.g., biofilm matrix-degrading enzymes. Alternatively mature biofilms can be perturbed by the use of dispersing agents (3), e.g., nitric oxide.
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Therapeutics 2015, 40, 277–283.
- O’Neill, J. Review on Antimicrobial Resistance. December 2014. Available online: http://amr-review.org/ (accessed on 15 September 2021).
- Carmen, J.C.; Roeder, B.L.; Nelson, J.L.; Ogilvie, R.L.R.; Robison, R.A.; Schaalje, G.B.; Pitt, W.G. Treatment of biofilm infections on implants with low-frequency ultrasound and antibiotics. Am. J. Infect. Control. 2005, 33, 78–82.
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561.
- Malik, A.; Mohammad, Z.; Ahmad, J. The diabetic foot infections: Biofilms and antimicrobial resistance. Diabetes Metab. Syndr. 2013, 7, 101–107.
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79.
- Wu, H.; Moser, C.; Wang, H.Z.; Høiby, N.; Song, Z.J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7.
- Pinto, S.N.; Dias, S.A.; Cruz, A.F.; Mil-Homens, D.; Fernandes, F.; Valle, J.; Andreu, D.; Prieto, M.; Castanho, M.; Coutinho, A.; et al. The mechanism of action of pepR, a viral-derived peptide, against Staphylococcus aureus biofilms. J. Antimicrob. Chemother. 2019, 74, 2617–2625.
- Ferreira, M.; Pinto, S.N.; Aires-da-Silva, F.; Bettencourt, A.; Aguiar, S.I.; Gaspar, M.M. Liposomes as a Nanoplatform to Improve the Delivery of Antibiotics into Staphylococcus aureus Biofilms. Pharmaceutics 2021, 13, 321.
- Jefferson, K.K.; Goldmann, D.A.; Pier, G.B. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 2005, 49, 2467–2473.
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39.
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320.
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881.
- Salmanoglu, E.; Kim, S.; Thakur, M.L. Currently Available Radiopharmaceuticals for Imaging Infection and the Holy Grail. Semin. Nucl. Med. 2018, 48, 86–99.
- Israel, O.; Keidar, Z. PET/CT imaging in infectious conditions. Ann. N. Y. Acad. Sci. 2011, 1228, 150–166.
- Eggleston, H.; Panizzi, P. Molecular imaging of bacterial infections in vivo: The discrimination of infection from inflammation. Informatics 2014, 1, 72–99.
- Ning, X.; Seo, W.; Lee, S.; Takemiya, K.; Rafi, M.; Feng, X.; Weiss, D.; Wang, X.; Williams, L.; Camp, V.M.; et al. PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew. Chem. Int. Ed. Engl. 2014, 53, 14096–14101.
- Auletta, S.; Varani, M.; Horvat, R.; Galli, F.; Signore, A.; Hess, S. PET Radiopharmaceuticals for Specific Bacteria Imaging: A Systematic Review. J. Clin. Med. 2019, 8, 197.
- Glaudemans, A.W.; Signore, A. FDG-PET/CT in infections: The imaging method of choice? Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 1986–1991.
- Erba, P.A.; Bandera, F.; Sollini, M.; Tascini, C. The use of 18F-FDG-PET/CT in the diagnostic workup of CIED infections: Another perspective. J. Am. Coll. Cardiol. 2012, 60, 1435–1436.
- Gopal, S.; Berg, D.; Hagen, N.; Schriefer, E.M.; Stoll, R.; Goebel, W.; Kreft, J. Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose. PLoS ONE 2010, 5, e10349.
- Kujundzic, E.; Fonseca, A.C.; Evans, E.A.; Peterson, M.; Greenberg, A.R.; Hernandez, M. Ultrasonic monitoring of early-stage biofilm growth on polymeric surfaces. J. Microbiol. Methods 2007, 68, 458–467.
- Vaidya, K.; Osgood, R.; Ren, D.; Pichichero, M.E.; Helguera, M. Ultrasound imaging and characterization of biofilms based on wavelet de-noised radiofrequency data. Ultrasound Med. Biol. 2014, 40, 583–595.
- Calliada, F.; Campani, R.; Bottinelli, O.; Bozzini, A.; Sommaruga, M.G. Ultrasound contrast agents: Basic principles. Eur. J. Radiol. 1998, 27 (Suppl. 2), S157–S160.
- Unnikrishnan, S.; Klibanov, A.L. Microbubbles as ultrasound contrast agents for molecular imaging: Preparation and application. AJR Am. J. Roentgenol. 2012, 199, 292–299.
- Anastasiadis, P.; Mojica, K.D.; Allen, J.S.; Matter, M.L. Detection and quantification of bacterial biofilms combining high-frequency acoustic microscopy and targeted lipid microparticles. J. Nanobiotechnol. 2014, 12, 24.
- Di Domenico, E.G.; Rimoldi, S.G.; Cavallo, I.; D’Agosto, G.; Trento, E.; Cagnoni, G.; Palazzin, A.; Pagani, C.; Romeri, F.; De Vecchi, E.; et al. Microbial biofilm correlates with an increased antibiotic tolerance and poor therapeutic outcome in infective endocarditis. BMC Microbiol. 2019, 19, 228.
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554.
- Bossù, M.; Selan, L.; Artini, M.; Relucenti, M.; Familiari, G.; Papa, R.; Vrenna, G.; Spigaglia, P.; Barbanti, F.; Salucci, A.; et al. Characterization of Scardovia wiggsiae Biofilm by Original Scanning Electron Microscopy Protocol. Microorganisms 2020, 8, 807.
- Asahi, Y.; Miura, J.; Tsuda, T.; Kuwabata, S.; Tsunashima, K.; Noiri, Y.; Sakata, T.; Ebisu, S.; Hayashi, M. Simple observation of Streptococcus mutans biofilm by scanning electron microscopy using ionic liquids. AMB Express 2015, 5, 6.
- Gomes, L.C.; Mergulhão, F.J. SEM Analysis of Surface Impact on Biofilm Antibiotic Treatment. Scanning 2017, 2017, 2960194.
- Ruan, X.; Deng, X.; Tan, M.; Yu, C.; Zhang, M.; Sun, Y.; Jiang, N. In vitro antibiofilm activity of resveratrol against avian pathogenic Escherichia coli. BMC Vet. Res. 2021, 17, 249.
- Alves, M.M.; Bouchami, O.; Tavares, A.; Córdoba, L.; Santos, C.F.; Miragaia, M.; de Fátima Montemor, M. New Insights into Antibiofilm Effect of a Nanosized ZnO Coating against the Pathogenic Methicillin Resistant Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 28157–28167.
- Franklin, M.J.; Chang, C.; Akiyama, T.; Bothner, B. New Technologies for Studying Biofilms. Microbiol. Spectr. 2015, 3, 1–32.
- Paddock, S.W. Principles and practices of laser scanning confocal microscopy. Mol. Biotechnol. 2000, 16, 127–149.
- Bayguinov, P.O.; Oakley, D.M.; Shih, C.C.; Geanon, D.J.; Joens, M.S.; Fitzpatrick, J.A.J. Modern Laser Scanning Confocal Microscopy. Curr. Protoc. Cytom. 2018, 85, e39.
- Neu, T.R.; Swerhone, G.D.W.; Lawrence, J.R. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 2001, 147, 299–313.
- Skogman, M.E.; Vuorela, P.M.; Fallarero, A. Combining biofilm matrix measurements with biomass and viability assays in susceptibility assessments of antimicrobials against Staphylococcus aureus biofilms. J. Antibiot. 2012, 65, 453–459.
- Strathmann, M.; Wingender, J.; Flemming, H.C. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Methods 2002, 50, 237–248.
- Okshevsky, M.; Meyer, R.L. Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms. J. Microbiol. Methods 2014, 105, 102–104.
- Kwon, H.Y.; Kim, J.Y.; Liu, X.; Lee, J.Y.; Yam, J.K.H.; Dahl Hultqvist, L.; Xu, W.; Rybtke, M.; Tolker-Nielsen, T.; Heo, W.; et al. Visualizing biofilm by targeting eDNA with long wavelength probe CDr15. Biomater. Sci. 2019, 7, 3594–3598.
- Kim, J.Y.; Sahu, S.; Yau, Y.H.; Wang, X.; Shochat, S.G.; Nielsen, P.H.; Dueholm, M.S.; Otzen, D.E.; Lee, J.; Delos Santos, M.M.; et al. Detection of Pathogenic Biofilms with Bacterial Amyloid Targeting Fluorescent Probe, CDy11. J. Am. Chem. Soc. 2016, 138, 402–407.
- Kwon, H.Y.; Kim, J.Y.; Lee, J.Y.; Yam, J.K.H.; Hultqvist, L.D.; Xu, W.; Rybtke, M.; Tolker-Nielsen, T.; Kim, J.J.; Kang, N.Y.; et al. CDy14: A novel biofilm probe targeting exopolysaccharide Psl. Chem. Commun. 2018, 54, 11865–11868.
- Ritenberg, M.; Nandi, S.; Kolusheva, S.; Dandela, R.; Meijler, M.M.; Jelinek, R. Imaging Pseudomonas aeruginosa Biofilm Extracellular Polymer Scaffolds with Amphiphilic Carbon Dots. ACS Chem. Biol. 2016, 11, 1265–1270.
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348.
- Daims, H.; Wagner, M. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 2007, 75, 237–248.
- Cerqueira, L.; Azevedo, N.F.; Almeida, C.; Jardim, T.; Keevil, C.W.; Vieira, M.J. DNA mimics for the rapid identification of microorganisms by fluorescence in situ hybridization (FISH). Int. J. Mol. Sci. 2008, 9, 1944–1960.
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS ONE 2011, 6, e14786.
- Malic, S.; Hill, K.E.; Hayes, A.; Percival, S.L.; Thomas, D.W.; Williams, D.W. Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiology 2009, 155, 2603–2611.
- Kaur, R.P.; Ludhiadch, A.; Munshi, A. Chapter 9—Single-Cell Genomics: Technology and Applications. In Single-Cell Omics; Barh, D., Azevedo, V., Eds.; Academic Press: Waltham, MA, USA, 2019; pp. 179–197.
- Alonso, A. DNA Extraction and Quantification. In Encyclopedia of Forensic Sciences, 2nd ed.; Siegel, J.A., Saukko, P.J., Houck, M.M., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 214–218.
- Larochelle, S. STOMPing at the bits. Nat. Methods 2015, 12, 1114.
- Domazet, B.; Maclennan, G.T.; Lopez-Beltran, A.; Montironi, R.; Cheng, L. Laser capture microdissection in the genomic and proteomic era: Targeting the genetic basis of cancer. Int. J. Clin. Exp. Pathol. 2008, 1, 475–488.
- Consentino, L.; Rejasse, A.; Crapart, N.; Bevilacqua, C.; Nielsen-LeRoux, C. Laser capture microdissection to study Bacillus cereus iron homeostasis gene expression during Galleria mellonella in vivo gut colonization. Virulence 2021, 12, 2104–2121.
- van Dam, G.M.; Themelis, G.; Crane, L.M.; Harlaar, N.J.; Pleijhuis, R.G.; Kelder, W.; Sarantopoulos, A.; de Jong, J.S.; Arts, H.J.; van der Zee, A.G.; et al. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-α targeting: First in-human results. Nat. Med. 2011, 17, 1315–1319.
- Kong, Y.; Yao, H.; Ren, H.; Subbian, S.; Cirillo, S.L.; Sacchettini, J.C.; Rao, J.; Cirillo, J.D. Imaging tuberculosis with endogenous beta-lactamase reporter enzyme fluorescence in live mice. Proc. Natl. Acad. Sci. USA 2010, 107, 12239–12244.
- Panizzi, P.; Nahrendorf, M.; Figueiredo, J.L.; Panizzi, J.; Marinelli, B.; Iwamoto, Y.; Keliher, E.; Maddur, A.A.; Waterman, P.; Kroh, H.K.; et al. In vivo detection of Staphylococcus aureus endocarditis by targeting pathogen-specific prothrombin activation. Nat. Med. 2011, 17, 1142–1146.
- van Oosten, M.; Schäfer, T.; Gazendam, J.A.; Ohlsen, K.; Tsompanidou, E.; de Goffau, M.C.; Harmsen, H.J.; Crane, L.M.; Lim, E.; Francis, K.P.; et al. Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 2013, 4, 2584.
- Bardhan, N.M.; Ghosh, D.; Belcher, A.M. Carbon nanotubes as in vivo bacterial probes. Nat. Commun. 2014, 5, 4918.
- Sauer, K. The genomics and proteomics of biofilm formation. Genome Biol. 2003, 4, 219.
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350.
- Hultin-Rosenberg, L.; Forshed, J.; Branca, R.M.; Lehtiö, J.; Johansson, H.J. Defining, comparing, and improving iTRAQ quantification in mass spectrometry proteomics data. Mol. Cell. Proteom. 2013, 12, 2021–2031.
- Luczak, M.; Formanowicz, D.; Marczak, Ł.; Suszyńska-Zajczyk, J.; Pawliczak, E.; Wanic-Kossowska, M.; Stobiecki, M. iTRAQ-based proteomic analysis of plasma reveals abnormalities in lipid metabolism proteins in chronic kidney disease-related atherosclerosis. Sci. Rep. 2016, 6, 32511.
- Trinh, H.V.; Grossmann, J.; Gehrig, P.; Roschitzki, B.; Schlapbach, R.; Greber, U.F.; Hemmi, S. iTRAQ-Based and Label-Free Proteomics Approaches for Studies of Human Adenovirus Infections. Int. J. Proteom. 2013, 2013, 581862.
- Suriyanarayanan, T.; Qingsong, L.; Kwang, L.T.; Mun, L.Y.; Truong, T.; Seneviratne, C.J. Quantitative Proteomics of Strong and Weak Biofilm Formers of Enterococcus faecalis Reveals Novel Regulators of Biofilm Formation. Mol. Cell. Proteom. 2018, 17, 643–654.
- Wang, K.; Wang, Y.; Wang, X.; Ren, Q.; Han, S.; Ding, L.; Li, Z.; Zhou, X.; Li, W.; Zhang, L. Comparative salivary proteomics analysis of children with and without dental caries using the iTRAQ/MRM approach. J. Transl. Med. 2018, 16, 11.
- Pham, T.K.; Roy, S.; Noirel, J.; Douglas, I.; Wright, P.C.; Stafford, G.P. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 2010, 10, 3130–3141.
- Zhang, F. Application of machine learning in CT images and X-rays of COVID-19 pneumonia. Medicine 2021, 100, e26855.
- Dimauro, G.; Deperte, F.; Maglietta, R.; Bove, M.; La Gioia, F.; Renò, V.; Simone, L.; Gelardi, M. A Novel Approach for Biofilm Detection Based on a Convolutional Neural Network. Electronics 2020, 9, 881.
- Xu, Y.; Li, C.; Jiang, Y.; Guo, M.; Yang, Y.; Yang, Y.; Yu, H. Electrochemical Impedance Spectroscopic Detection of E.coli with Machine Learning. J. Electrochem. Soc. 2020, 167, 047508.
- Buetti-Dinh, A.; Galli, V.; Bellenberg, S.; Ilie, O.; Herold, M.; Christel, S.; Boretska, M.; Pivkin, I.V.; Wilmes, P.; Sand, W.; et al. Deep neural networks outperform human expert’s capacity in characterizing bioleaching bacterial biofilm composition. Biotechnol. Rep. 2019, 22, e00321.
- Chudzik, P.; Majumdar, S.; Calivá, F.; Al-Diri, B.; Hunter, A. Microaneurysm detection using fully convolutional neural networks. Comput. Methods Programs Biomed. 2018, 158, 185–192.
- Guo, Y.; Budak, Ü.; Şengür, A. A novel retinal vessel detection approach based on multiple deep convolution neural networks. Comput. Methods Programs Biomed. 2018, 167, 43–48.
- Gelardi, M.; Passalacqua, G.; Fiorella, M.L.; Mosca, A.; Quaranta, N. Nasal cytology: The “infectious spot”, an expression of a morphological-chromatic biofilm. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1105–1109.
- Wong, P.-C.; Fan, T.-E.; Lee, Y.-L.; Lai, C.-Y.; Wu, J.-L.; Chang, L.-H.; Su, T.-Y. Detection and Identification of the Stages of DH5-Alpha Escherichia coli Biofilm Formation on Metal by Using an Artificial Intelligence System. Microscopy Microanal. 2021, 27, 1218–1225.
- Pobre, V.; Arraiano, C.M. Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli. BMC Genom. 2015, 16, 72.
- Saramago, M.; Domingues, S.; Viegas, S.C.; Arraiano, C.M. Biofilm formation and antibiotic resistance in Salmonella Typhimurium are affected by different ribonucleases. J. Microbiol. Biotechnol. 2014, 24, 8–12.
- Cohen, D.; Mechold, U.; Nevenzal, H.; Yarmiyhu, Y.; Randall, T.E.; Bay, D.C.; Rich, J.D.; Parsek, M.R.; Kaever, V.; Harrison, J.J.; et al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 11359–11364.
- Lehnik-Habrink, M.; Schaffer, M.; Mäder, U.; Diethmaier, C.; Herzberg, C.; Stülke, J. RNA processing in Bacillus subtilis: Identification of targets of the essential RNase Y. Mol. Microbiol. 2011, 81, 1459–1473.
- Baumgardt, K.; Charoenpanich, P.; McIntosh, M.; Schikora, A.; Stein, E.; Thalmann, S.; Kogel, K.H.; Klug, G.; Becker, A.; Evguenieva-Hackenberg, E. RNase E affects the expression of the acyl-homoserine lactone synthase gene sinI in Sinorhizobium meliloti. J. Bacteriol. 2014, 196, 1435–1447.
- Moreira, R.N.; Dressaire, C.; Barahona, S.; Galego, L.; Kaever, V.; Jenal, U.; Arraiano, C.M. BolA Is Required for the Accurate Regulation of c-di-GMP, a Central Player in Biofilm Formation. mBio 2017, 8, e00443-17.
- Obana, N.; Nakamura, K.; Nomura, N. Role of RNase Y in Clostridium perfringens mRNA Decay and Processing. J. Bacteriol. 2017, 199, e00703-16.
- Diallo, I.; Provost, P. RNA-Sequencing Analyses of Small Bacterial RNAs and their Emergence as Virulence Factors in Host-Pathogen Interactions. Int. J. Mol. Sci. 2020, 21, 1627.
- Chakravarty, S.; Massé, E. RNA-Dependent Regulation of Virulence in Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 337.
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251.
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganisms. Heliyon 2019, 5, e02192.
- Sharma, A.; Gupta, P.; Kumar, R.; Bhardwaj, A. dPABBs: A Novel in silico Approach for Predicting and Designing Anti-biofilm Peptides. Sci. Rep. 2016, 6, 21839.
- Porto, W.F.; Irazazabal, L.; Alves, E.S.F.; Ribeiro, S.M.; Matos, C.O.; Pires, Á.S.; Fensterseifer, I.C.M.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 2018, 9, 1490.
- Di Luca, M.; Maccari, G.; Maisetta, G.; Batoni, G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015, 31, 193–199.
- Gupta, S.; Sharma, A.K.; Jaiswal, S.K.; Sharma, V.K. Prediction of Biofilm Inhibiting Peptides: An In silico Approach. Front. Microbiol. 2016, 7, 949.
- Rajput, A.; Thakur, A.; Sharma, S.; Kumar, M. aBiofilm: A resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res. 2018, 46, D894–D900.
- Srivastava, G.N.; Malwe, A.S.; Sharma, A.K.; Shastri, V.; Hibare, K.; Sharma, V.K. Molib: A machine learning based classification tool for the prediction of biofilm inhibitory molecules. Genomics 2020, 112, 2823–2832.
More
