Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Sung Un Huh and Version 2 by Dean Liu.
RNA binding proteins (RBPs) have been known to control target genes. One type of protein, Pumilio (Pum)/Puf family RNA binding proteins, show a specific binding of 3′ untranslational region (3′ UTR) of target mRNA and function as a post-transcriptional/translational regulator in eukaryotic cells. Plant Pum protein is involved in development and biotic/abiotic stresses.
- RNA binding protein
- Pumilio
- translational modification
- RNA biomolecules
1. Protein–Protein Interaction in Plant Pum
Pum-HD has a dual function. The inner surface of Pum-HD binds to the conserved ‘UGUX3-5UA’ motif in the 3′ UTR regions of the target gene. Interestingly, the outer surface of Pum-HD also permits protein–protein interactions with diverse proteins [1][2][6,41]. The yeast Pum protein, PUF5p, binds to and represses HO mRNA, which encodes a DNA endonuclease via binding to the Pumilio-RNA binding motif. Pop2p, which encodes a component of the Ccr4p-Pop2p-Not deadenylase complex, is required for PUF5p-mediated HO mRNA decay [3][42]. It affects both mRNA stability and translational efficiency [4][5][43,44]. Thus, Pums do not work alone to repress the target mRNA. Furthermore, yeast PUF5p and Pop2p interaction is evolutionally conserved in C. elegans PUF8 and H. sapiens (PUM1) [3][42]. Arabidopsis Pum, APUM5, also interacts with plant Ccr4 homologues [6][45]. Sometimes, yeast PUF5p does not require Ccr4-Pop2 deadenylase when PUF5p responds to DNA replication stress [7][46]. Thus, plant Pum function could be associated with deadenylase-dependent and -independent pathways via formation, as with Ccr4-Pop2-NOT mRNA deadenylase complexes.
Human PUM2 can make a complex with Argonaute (Ago) miRNA-binding protein, and then, the PUM2-Ago heterodimer is associated with the core translational elongation factor (eEF1A). This complex inhibits eEF1A GTPase activity and translational elongation [8][47]. These results imply that plant Pum could be associated with the Ago-eEF1A protein complex.
2. The Role of Pum in Plant Development
Pum proteins play important roles during development, differentiation and cell cycle regulation in various organisms [9][10][33,50]. APUM1 to APUM6 are specifically associated with shoot stem cell maintenance genes such as WUSCHEL (WUS) and CLAVATA1 (CLV1) [11][12][13][34,51,52]. Like the mammalian system, human PUM2 is expressed in embryonic stem cells and could affect germ cell development [14][53]. Mammalian PUM1 and PUM2 regulate cell cycle inhibitor Cdkn1b via translational control [15][54]. C. elegans Pum, PUF8, might function as a repressor of the stem cell proliferative fate via control of GLP-1/Notch signaling in germline cells [16][55]. Likewise, plant Pum could affect a variety of development stages. Gene expressions of APUM23 and APUM24 were shown to be continuously expressed in all the developmental stages and were enhanced at the seed stage (Figure 2A). APUM23 affects pre-ribosomal RNA processing in the nucleolus and the apum23 mutant showed a developmental defeat phenotype [17][37]. It is possible that APUM24 could function in the ribosomal RNA processing because APUM24 predominantly localizes to the nucleolus [18][49]. APUM5 and APUM6 were highly abundant at developmental stages (Figure 3). However, these mutants did not show abnormal developmental phenotypes. APUM1 to APUM6 have a similar protein structure and might have the same redundancy. In the future, this will be studied for genetic crossover with APUM1 to APUM6 mutants. In rice, at least four rice Pum genes are highly expressed at the vegetative stage (Figure 2B). Plant Pum proteins could play a role in development and differentiation.
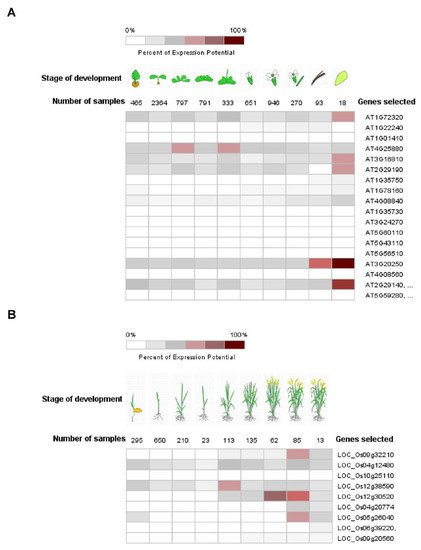
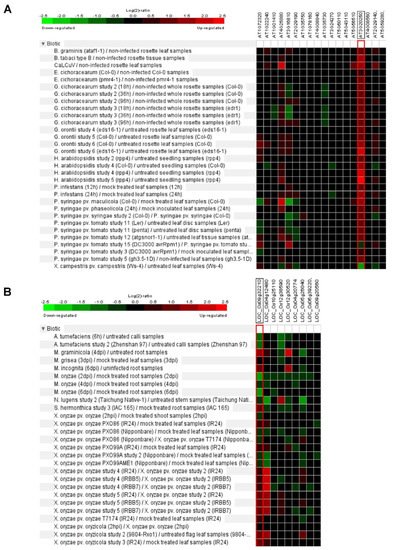

Figure 2. Expression pattern of Arabidopsis and rice Pum in development. (A,B) Expression pattern of Arabidopsis and rice Pum was analyzed at developmental stage using the Genevestigator (https://www.genevestigator.com accessed on 13 November 2021). Expression levels of Arabidopsis and rice Pum vary under developmental stages. Transcript levels were assessed using microarray data available from Genevestigator.

Figure 3. Biotic stress response of Arabidopsis and rice Pum.
3. Plant Pum Function in Biotic Stress
In Arabidopsis, some putative target RNAs of Pum were identified by yeast in a three-hybrid system. These genes contained Pumilio RNA binding motifs at the 3′ UTR regions [11][34]. For example, Responsive to dehydration 19 (RD19) and Plant homeodomain (PHD) containing protein (At3g63500) were isolated [11][34]. RD19 encodes cysteine protease and is involved in resistance and gene-mediated immunity via interaction with Ralstonia solanacearum type III effector PopP2 [19][56]. This means that plant Pum could be related to plant immunity. APUM5 is involved in plant defense against CMV infection and function as a translational repressor via binding to the 3′ UTR of CMV [20][32]. Furthermore, gene expression analysis of Arabidopsis Pum family exhibits enhanced or repressed expression levels upon the Cabbage leaf curl virus (CaLCuV), Golovinomyces cichoracearum, Hyaloperonospora arabidopsidis, Peudomonas syringae and Xanthomonas campestris (Figure 3A). Similarly, rice Pum genes are upregulated or downregulated by Mycosphaerella graminicola, Meloidogyne incognita, Magnaporthe oryzae, Nilaparvata lugens and Xanthomonas oryzae infections (Figure 3B). When infected with M. oryzae, most of the rice Pum genes were shown to be repressed. Expression levels of OS09g32210 and OS04g12480 genes were highly enhanced upon X. oryzae infection in IRBB5/7 (resistance) and IR24 (susceptible) rice plants (Figure 3B). Thus, infectious pathogens may affect plant Pum gene expression and attenuate plant immunity. In D. melanogaster Pum mutants, some antibacterial genes are highly expressed [21][19]. In particular, CG18372 (Attactin-B), CG4740 (Attactin-C), CG7629 (Attactin-D), CG1373 (Cecropin C) and CG13422 (Gram-negative binding protein) were encoded antibacterial peptides [21][19]. The immunity function of Pum in plant and mammalian genes will be important to investigate in the extended study of transcriptional/translational control.
4. Plant Pum Function in Abiotic Stress
Phytohormone abscisic acid (ABA) and brassinosteroid (BR) have essential roles in the manipulation of abiotic stress responses [22][23][57,58]. Expression patterns of Arabidopsis Pum are dynamically changed by ABA and BR treatment (Figure 4A). Recently, the apum23 mutant exhibited altered gene expression patterns of ABA and salt-stress-responsive genes. APUM23 is essential for salt sensitivity in response salt stress [24][59]. APUM5 has also been reported to be involved in salt tolerance [25][60]. The overexpression of APUM9 results in enhanced heat stress tolerance [26][61]. Furthermore, the expression of several Arabidopsis Pum genes is changed by environment stresses such as cold, drought and salt stresses (Figure 4A). Rice Pum genes responded to auxin and cytokinin but not ABA (Figure 4B). Basically, cytokinin and auxin are also deeply associated with abiotic stress responses [27][28][62,63]. Plant Pum can influence the target RNA through environmental stresses and phytohormone molecules.
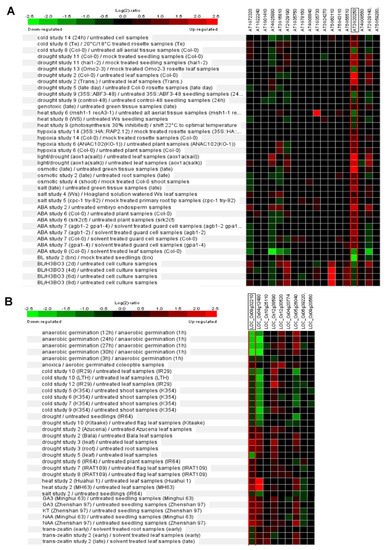

Figure 4. Differential expression patterns of Pum genes in Arabidopsis and rice in response to abiotic stresses or phytohormones. (A) Expression pattern of Arabidopsis Pum was analyzed in abiotic stresses (cold, drought, heat, hypoxia, osmotic and salt) and phytohormones (ABA and brassinosteroid) treatments using Genevestigator. (B) Expression pattern of rice Pum was analyzed in abiotic stresses (anaerobic, cold, drought, heat, and salt) and phytohormones (gibberellin, auxin, and cytokinin) treatments using Genevestigator. Arabidopsis and rice Pum transcript levels were assessed using microarray data available from Genevestigator. Expressions patterns were searched in a range of abiotic stress conditions in this database. Red colors indicate genes that were upregulated relative to the control for a given treatment, and green colors indicate genes that were downregulated relative to the control.
5. The Multifunctional Plant Pum Protein and Crop Engineering
The translational control of gene expression is an important step for the survival of plants. Pum proteins evolutionally conserve the canonical Pum-HD RNA binding domain in most of the eukaryotes. This Pum-HD confers target RNA specificity via direct binding and functions as a protein–protein interaction platform. Sometimes, Pum acts as a translational activator. Until now, plant Pum functions were not well known. The plant Pum family contains the highest number of members, about twenty, compared with the mammalian Pum family. This implies that many plant Pum proteins might have plant-specific functions and could have plant-specific targets. From the in silico data analysis, it is shown that plant Pums could be associated with a variety of signaling pathways. This regulation could be more complicated because plant hormones exhibit crosstalk between different plant hormones. A better understanding of the plant Pum regulation mechanism would help innovate new strategies to improve crop plant engineering. Thus, it would be interesting to identify the putative RNA targets and generate customized Pum-HD proteins for agriculture.
