Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by marta baiocchi and Version 2 by Vicky Zhou.
Colorectal cancer (CRC) represents one of the most deadly cancers worldwide. Colorectal cancer stem cells (cCSCs) are the driving units of CRC initiation and development. After the concept of cCSC was first formulated in 2007, a huge bulk of research has contributed to expanding its definition, from a cell subpopulation defined by a fixed phenotype in a plastic entity modulated by complex interactions with the tumor microenvironment, in which cell position and niche-driven signals hold a prominent role. The wide development of cellular and molecular technologies recent years has been a main driver of advancements in cCSCs research
- cancer stem cells
- colorectal cancer
- cancer stem cell methods
1. Introduction
According to the stem cell model, most tumors, including colorectal cancer (CRC), contain a small population of cancer stem cells (CSCs) that are deeply implicated in tumor generation and progression, drug resistance, recurrence, and metastasis [1][2][1,2]. The characterization of the molecular and functional features of colorectal CSCs (cCSCs) has thus received intense research efforts in recent years due to its promise to reveal new routes of intervention for tumor and metastasis eradication. Methods and concepts for understanding CSCs biology in solid tumors historically derived from studies on normal hematopoiesis and leukemia. In fact, the CSC concept itself originated from landmark research [3][4][3,4] that in the early nineties first showed that leukemia is organized as a hierarchical system, mimicking that of the normal hematopoietic system. The neoplastic cell hierarchy in leukemias develops from a small subset of stem cells able to both self-renew and to give rise to a cascade of more differentiated cells. Notably, the golden standard for the identification of leukemic stem cells (LSCs) was established by extending the definition for normal hemopoietic cells as cells functionally capable of initiating neoplasia into recipient mice. A few years later, the experimental methods that led to LSCs identification were translated to solid tumors in general, and in particular to CRC. During the following years, the expanding range of methodological approaches applied to cCSCs biology has paralleled a continuous evolution of the cCSCs concept, providing a significant example of knowledge advancement about a complex biological issue. The main technical approaches that contributed to cCSCs identification and characterization, in particular of human CRC, are summarized in Figure 1, which also underlines the high degree of intersection of their application. Methods for cCSCs identification can be broadly summarized as (1) cell isolation by fluorescence-activated cell sorting, (2) cell culture-based selection systems, (3) transplantation into recipient animals, and (4) lineage tracing techniques. Here, we will revise the major advancements that led to the development of the current cCSC concept, keeping a historical view on the evolution of technologies that allowed cCSC characterization to the present day (Figure 2).
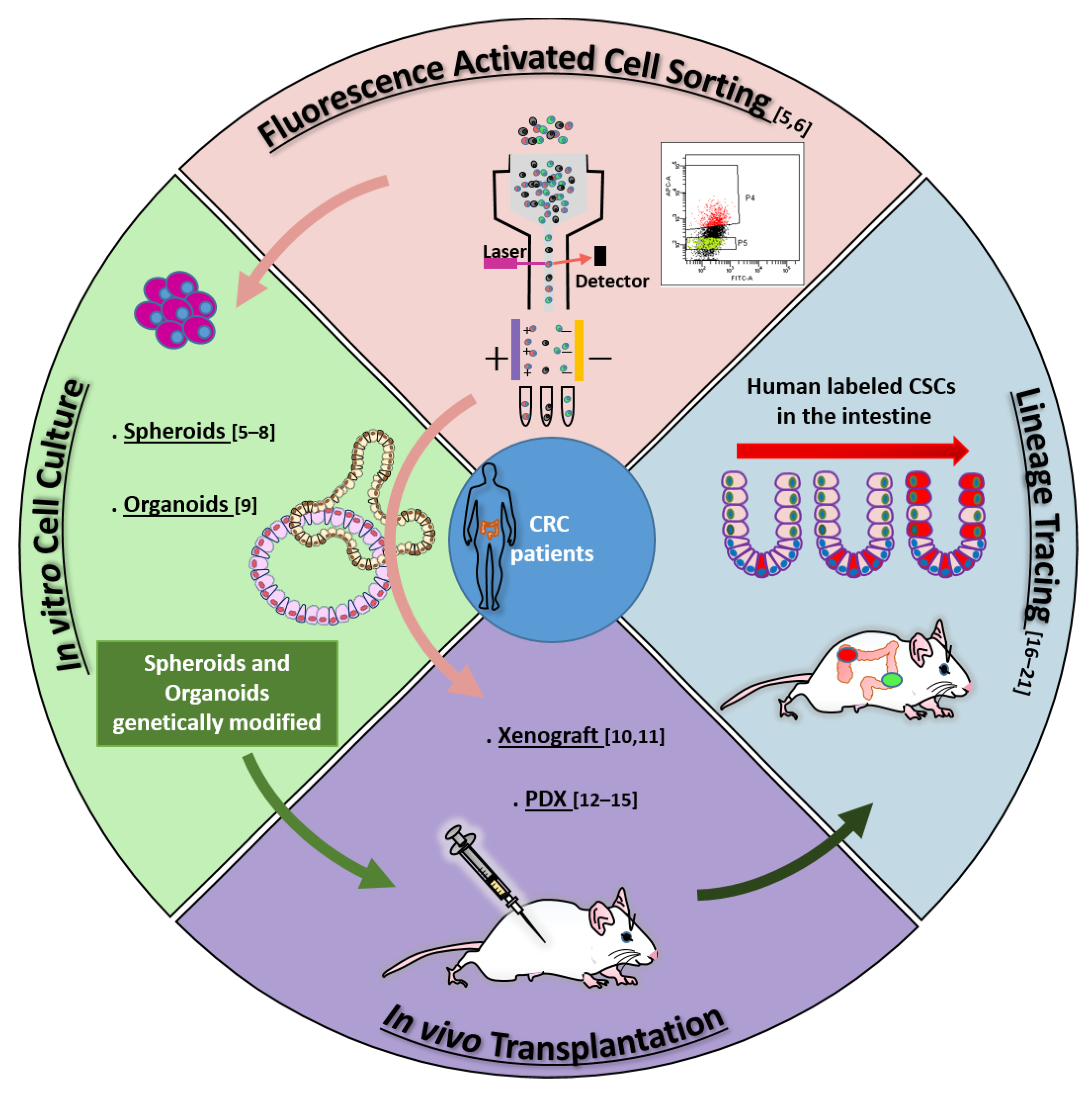
Figure 1. Main methodologies contributing to cCSCs definition: fluorescence-activated cell sorting (FACS) [5][6][5,6]; in vitro cell culture [5][6][7][8][9][5,6,7,8,9]; in vivo transplantation [10][11][12][13][14][15][10,11,12,13,14,15]; lineage tracing [16][17][18][19][20][21][16,17,18,19,20,21]. Arrows across sectors identify the complementary application of different techniques: For example, identification of cCSCs by cell sorting can be validated by in vitro cell culture and/or by in vivo transplantation assays (pink arrows); Xenografting is often performed with cultured and/or genetically labeled cCSCs (light green arrow); human cCSC lineage tracing is mostly performed by transplanting cCSCs previously genetically manipulated in culture (dark green arrow). Numbers in parentheses indicate references.
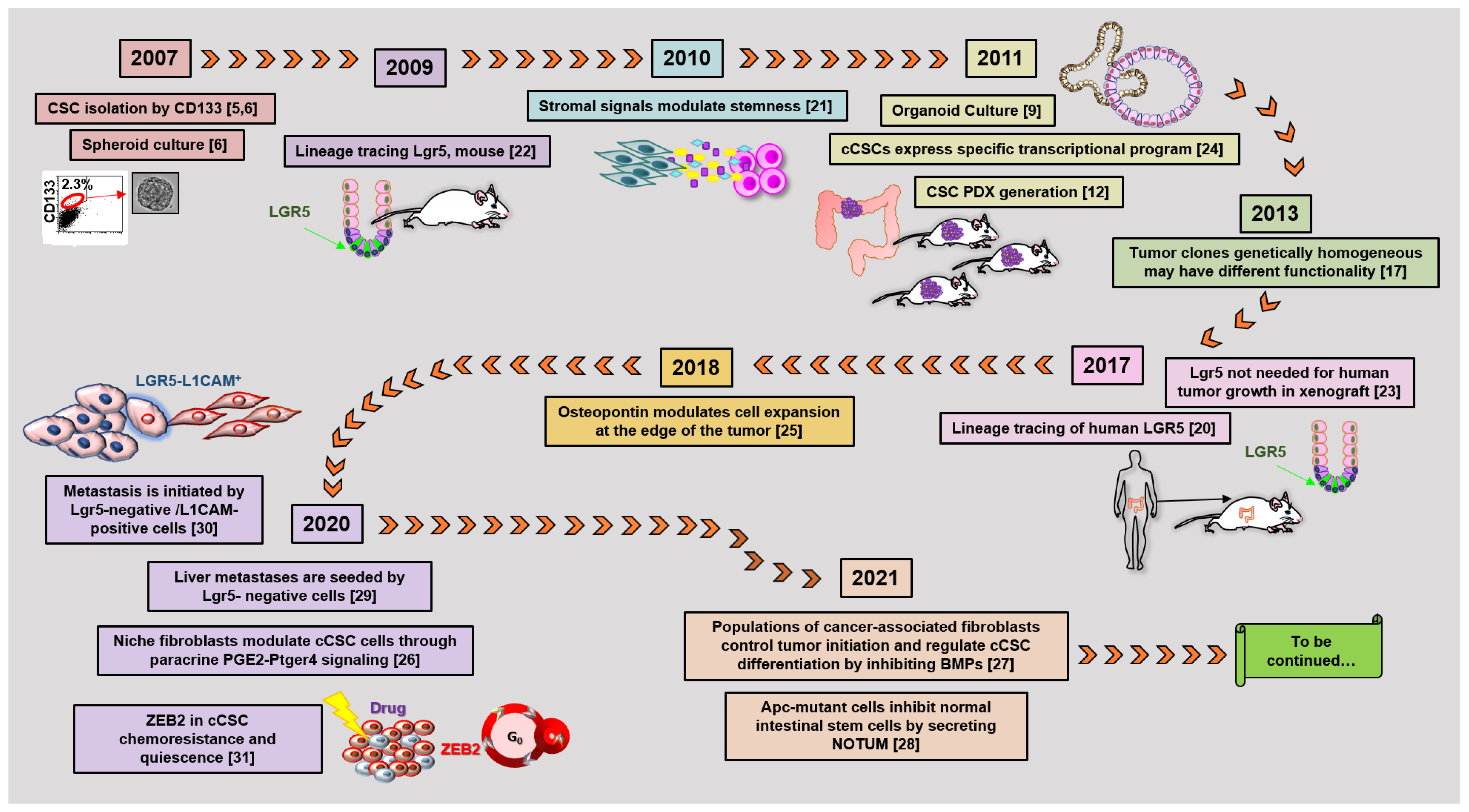
Figure 2. The timetable of landmark studies that have contributed to defining cCSC biological features and functions. Numbers in parentheses indicate references [5][6][9][12][17][20][21][22][23][24][25][26][27][28][29][30][31][5,6,9,12,17,20,21,22,23,24,25,26,27,28,29,30,31].
2. Merging Methodologies and Evolving Concepts: cCSC Plasticity and the Niche
The different approaches discussed above have altogether contributed to defining cCSC as a cell subpopulation driving initiation, development, and growth of CRC. Nevertheless, parallel research has challenged the concept that a fixed phenotype could be attributed to such population(s). Early observations indeed indicated that cells not expressing specific markers might undertake a functional stem cell role, at least under stress circumstances. Among these, Shmelkov et al. followed intestinal murine CD133 expression by a Lac-Z reporter showing that both CD133+ and CD133– cells can initiate metastases into immunodeficient mice [32][77]. Later, Lgr5+ cells were found dispensable for adenoma formation after irradiation in Apc-mice [33][78], while a population of keratin-19 (KRT19)-positive, Lgr5-negative cells were shown to be responsible for cancer tissue regrowth following irradiation in mice [34][79]. More recently, de Sousa et al. analyzed the effects of Lgr5+ cell removal through an inducible diphtheria toxin in a model of murine colon cancer. After killing Lgr5+ cells by toxin activation, tumor growth was not impaired, indicating that other cell type(s) could take over a stem cell function [35][80]. Similar results were shown in human CSCs, by xenotransplanting organoids carrying an inducible gene for caspase 9 inserted into the Lgr5 locus. Elimination of Lgr5+ cells by caspase activation blocked tumor growth. Upon removal of the killing stimulus, however, tumor regrowth was driven by a population of Lgr5−/KRT20 positive cells, which regenerated Lgr5+ cells [23]. The observations that stem cell marker-expressing cells may not be the unique drivers of adenoma/carcinoma points to the issue of stem cell plasticity. This concept includes both the capability of intestinal CSCs to acquire different phenotypes and the potential of different tumor cell subpopulations to take on stem cells function under different circumstances [1][2][36][1,2,81]. A scheme comparing the hierarchical model of cCSC versus the emerging model of plastic cCSC is shown in Figure 3.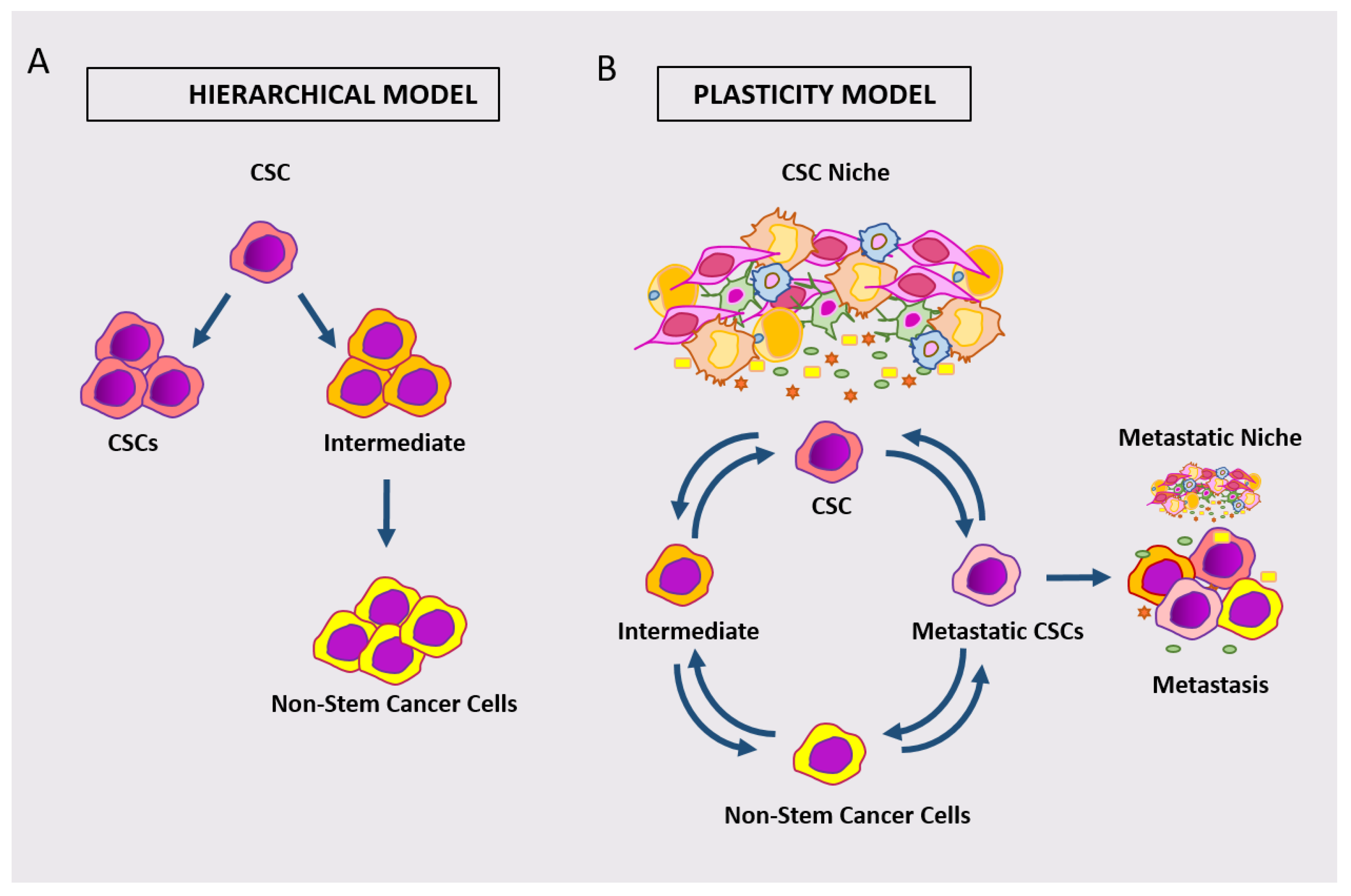
Figure 3. Schematic representation of the hierarchical model of cCSC (A) versus the model of plastic cCSC (B). According to the plastic model, different classes of cancer cells can dynamically take on stem cell phenotypes/functions.
The plasticity of cCSCs was also supported by other studies tracking stem cells on the base of Wnt activation state rather than through stem cell marker expression. In fact, tracing human xenografted cCSCs by means of a Wnt-dependent GFP reporter showed that cCSCs driving tumor expansion are located at the edge of the tumor. In this system, signals from surrounding stromal cells proved to be instrumental in modulating Wnt activation levels in tumor cells, pointing to a role for stroma in inducing cCSC function [21]. In turn, constitutive NF-kB activation was reported to enhance Wnt activation and stem cell marker expression in mouse crypt cells [37][82]. More recent phenotype-independent tracking experiments keep extending this concept. Phenotype-independent lineage tracing of human cCSCs confirmed that clonal expansion in colon cancer xenografts mostly arises at the leading edge and that cell position is a main driver of clonal competition during tumor development [38][83]. Other studies used an improved multicolor marker-independent tracing system (RGB/LeGO) to follow long-term clonal dynamics within xenografts, showing a correlation between clone size and proximity to the edge. These observations further support a persisting role of tumor geometry and cell position in orchestrating clonal competition during tumor growth [39][84].
The plastic nature of intestinal CSCs is also consistent with the knowledge that genetically homogeneous cells within the tumor can take over different functions, as demonstrated by clonal analysis of expanding human cCSC clones in xenografts by lentiviral marking [17] and by ultra-deep whole-genome sequencing tracking SNV/CNV [18]. In addition, cCSCs are known to express specific transcriptional programs [24][40][41][24,85,86], and a recent report has shown that high levels of ribosomal activity and protein synthesis individuate cCSCs independently from their specific mutational landscape [42][87]. It is noteworthy that genetic studies on patient’s adenoma and CRC tissues collectively indicate that driver mutations are mostly established early in the first stages of tumorigenesis, while limited functional mutational divergence is added during tumor development, thus strengthening the idea that non-genetic events are the main determinants of intestinal CSCs function during tumor development [43][44][45][88,89,90].
The concept that cCSC plasticity may be related to cell location points to the role of the so-called tumor niche in instructing cCSC behavior. In fact, cells at the tumor’s edge reside in close proximity to stromal cells, thus being exposed to stroma-derived signals. Early studies had actually described an instructive role of the niche and of niche-secreted factors, including WNTs, R-spondins, and BMP-inhibitors, in influencing the fate of intestinal CSCs [37][46][47][48][49][82,91,92,93,94]. In recent years, research on tumor niche has been expanding, disclosing the complexity of the crosstalk that orchestrates the plastic features of cCSCs. In this frame, Lenos et al. recently described a relevant role of stroma-secreted osteopontin as an inducer of cell expansion at the edge of the tumor [25]. Another report identified a specific subpopulation of fibroblasts in the mesenchymal tumor niche, which controls tumor-initiating cells through paracrine PGE2 (Prostaglandin E2)-Ptger4 signaling [26]. In turn, polarized populations of cancer-associated fibroblasts regulate cCSC differentiation and cancer progression by balanced inhibition of BMPs by GREM1 [27].
In this frame, transient epigenetic modifications, including variation in DNA methylation, histone modification, and chromatin accessibility, certainly contribute to sustaining CRC cell stemness [50][51][52][95,96,97]. Several epigenetic mechanisms capable of affecting CRC cell stemness, including but not limited to Wnt pathway activation/inactivation, have been described [53][54][55][56][57][58][98,99,100,101,102,103]. In this context, cell position-related epigenetic modulation likely holds a particularly relevant role in cCSC plasticity [50][51][52][95,96,97]. For example, tumor cells located at the edge or in central areas of the tumor are exposed to varying concentrations of metabolites and oxygen, which can modulate histone modifications and DNA methylation [51][96]. Stroma-secreted factors acting on cCSC plasticity, among which TGFβ, in turn, are able to induce cell epigenetic modifications [59][104]. Novel methods for single-cell epigenetic analyses, aided by developing computational systems, are now beginning to shed light on inherited epigenetic states of cellular lineages within CRC [60][61][105,106].
Further complexity is added to the picture by the finding that cCSCs are able to deliver inhibitory signals to normal intestinal cells, both directly and by inducing stromal cells to secrete specific factors. In this frame, two studies recently described the capability of Apc-mutated adenoma cells to inhibit stem cell activity of non-mutated cells within the same crypt and adjacent crypts through secretion of soluble Wnt antagonists [62][107]. Among these factors, a prominent role is played by NOTUM, whose pharmacological inhibition blocks adenoma formation [28]. Other important information has been provided by an innovative tracking system, the Confetti-derived Red2Onco, by which oncogenes such as mutated KRAS or PI3K are inserted only in RFP+ tumor cells. By allowing tracking of separately normal and mutated intestinal cells, this system has revealed that oncogene-driven signals from mutated cells induce apoptosis and differentiation of surrounding normal intestinal stem cells, both directly and by instructing surrounding stromal cells to secrete inhibitory factors [63][108].
The plasticity of cCSCs holds a particular interest in view of their capability to initiate metastasis. Several recent studies have described variable stem cell marker expression in metastasis-initiating cCSCs. For example, in the study by de Sousa e Melo already mentioned, while the selective elimination of Lgr5+ cells did not lead to tumor growth stopping, metastasis initiation in the liver was delayed until the Lgr5+ cell re-emerged [35][80]. By using intravital microscopy on xenotransplanted organoids carrying an inducible Lgr5-EGFP-Confetti, Fumagalli et al. followed metastases seeding and initiation, observing that liver metastases are seeded by Lgr5− cells, although Lgr5 positivity and stem cell marker expression re-emerges in growing metastases [29]. Consistently, Ganesh et al. showed that patients’ metastases are initiated by cells overexpressing L1CAM+, that do not concurrently express Lgr5 [30]. Altogether, a picture thus emerges, in which Lgr5 expression is downregulated at some stages of dissemination/seeding, to be then re-expressed during metastasis growth into target organs. Low expression of stem cell markers of metastasis-initiating cCSCs is indeed shared by budding cells at the edge of the tumor [64][109] and by cCSCs circulating in the blood flow, i.e., putative migrating cells at metastasis target organs [65][66][67][68][110,111,112,113]. The plasticity of metastasis-initiating/circulating cCSCs has been put into relationship with the transition of cCSCs into quiescent/drug-resistant states [31][69][31,114], and involves at least at some stages EMT (epithelial to mesenchymal transition, reviewed in [67][112]). This capability of cCSCs to downregulate epithelial and/or stem cell markers to take on mesenchymal features is driven by specific transcription factors, including SNAI1, SNAI2, ZEB1, ZEB2, and TWIST1 [67][70][71][112,115,116]. The crosstalk between tumor and stromal cells holds a role of utmost importance in CRC and in cCSCs, in particular through TGFβ signaling [72][73][74][62,63,117]. Finally, other mechanisms of stromal/tumor cell modulating metastatic cCSCs are emerging: a recent report details the reciprocal reinforcement between visceral adipose stromal cells and metastatic CD44v6-positive cCSCs, sustained by adipose cell-secreted IL-6 and HGF, and by neurotropin produced by CD44v6+ cells [75][118].
3. Conclusions and Future Perspectives
Taken together, most recent studies have converged to redefine CRC cell stemness, from the permanent feature of a restricted tumor cell subpopulation to a function that can be undertaken by different cell types under different circumstances. Such a function is modulated by a series of factors awaiting further dissection, but definitely including signals both intrinsic and from the microenvironment [1][2][1,2]. Spatial constraints likely also contribute to influencing the clonal development of CRC [76][119]. It has been proposed that different parameters may dictate clone competition during different developmental phases of tumors, arguing that space constraints may be determinant in leukemia developing into the bone marrow, while stromal signals may have a prevalent role in solid tumors [52][97]. Even within the same tumor, different parameters may acquire or lose relevance during development: Regarding intestinal cancer, it is easy to hypothesize that cell clonogenicity and stemness may be differently regulated during the initial adenomatous phase, within the restricted crypt environment, and later on, at the expanding invasive edge of carcinoma. Several pieces of evidence indeed indicate that different clonal selection determinants act in adenoma as compared to carcinoma [77][78][120,121]. Altogether, the picture of intestinal CSCs represents a fast developing, challenging biological issue.
The wide array of methodological advancements of these years is increasing our knowledge of the cellular events taking place in CRC at a fast speed [79][122]. New cell culture methods are progressively extending into sophisticated engineering methodologies, among which biomimetic scaffolds and organs on chips [80][81][123,124]. Single-cell microfluidics are already contributing to the characterization of circulating cCSCs, and begin to find application in tissue-dissociated cells [52][97]. Other important developments of in vitro methods aim to reconstruct the contribution of tumor microenvironment components, including co-culture systems of organoids with mesenchymal cells [82][125] or lymphocytes [83][126]. Intravital microscopy allows visualizing cells within a whole organ [29][84][29,127]. Lineage tracing is increasingly taking advantage by developing high-throughput sequencing techniques [85][73], allowing unsupervised tagging by lentivirus or by CRISPR, or even by following naturally occurring mutations [86][128]. Advanced single-cell technologies [87][88][129,130], including genome sequencing [89][131] and scRNA analysis [90][91][132,133], are already contributing to colorectal CRC and niche studies [60][92][93][105,134,135] and are expected to gain further strength in the very near future [52][97].
As a final note, the growing impact of computational methods in research on cancer cell biology, and CSCs in particular, deserves a special mention. The analysis and management of the enormous amount of data generated by multi-omic techniques are in fact made possible only by the continuous development of dedicated algorithms, and most of the recent studies mentioned in this review have heavily taken advantage of increasingly sophisticated computational approaches (see for example [39][43][61][62][90][94][84,88,106,107,132,136]). Recent applications include meta-approaches able to integrate data derived from multiple different analytical tools, such as scDNA- and scRNA-sequencing [52][97]. Complex systems of lineage tracing reconstruction have been developed, such as the so-called pseudotime projection analyses, able to elaborate cell lineage developmental trajectories based on scRNA expression patterns [86][128]. Similarly, genetic lineage tracing by scDNA-seq allows the reconstruction of spatial models of clonal development in solid cancers [43][88][88,130].
Self-training artificial intelligence (AI) tools, in particular, machine learning (ML) and deep learning (DL), have demonstrated an exceptional power in individuating patterns within wide datasets and are currently not only applied to cancer development research but also evaluated for clinical classification and decision-making [95][96][137,138]. The specific capability of DL to discriminate and classify images, in particular, surpassed that of humans in 2005 [96][138], and it is now widely used to analyze large-size imaging datasets. Most importantly, it is also generating breakthrough innovative imaging technologies, among which label-free cell recognition systems, able to predict fluorescent labels in unlabeled microscopy images [97][139] or ghost cytometry, that allow sorting of cells on the base of marker-free, image-free cell morphology pattern analysis [98][140].
In conclusion, it is easy to prophesize that such a wide range of fast-developing methodologies are destined to enlighten an increasing complexity of the cCSCs model in the very near future.
