PGPR promotes plant growth indirectly by preventing phytopathogens by producing metabolites of antimicrobial nature; the production of enzymes such as chitinase, protease, and lipase, which enable lysis of pathogenic bacteria and fungi; and induction of systemic resistance. PGPR produces low molecular weight compounds possessing antimicrobial activity even at low concentrations. Due to these compounds, PGPRs are the first choice among biological control agents for sustainable agriculture.
- plant growth-promoting rhizobacteria
- siderophore
- stress resistance
- enzyme
- phytohormone
- induced systemic resistance
- gene expression
1. Introduction
- Introduction
Indiscriminate use of agrochemicals led to deterioration of soils’ biotic communities, widespread environmental contamination by agrochemical residues, and significant negative impacts on public health [1,2], while combustion of fossil fuels and emissions of greenhouse gases are accelerating global climate changes [3]. Global climate change leads to the generation of abiotic stresses such as drought, salinity, and temperature extremes, which directly influence plants and result in decreased productivity. Abiotic stresses perplex plant growth and development and delay seed germination and enzyme activities [4,5]. Abiotic stresses also hinder soil microbial diversity and physicochemical properties of soil, resulting in lower productivity and yield loss [6]. To counteract the negative impacts of stress on crop plants, the agricultural policy is accentuating sustainable production systems with an emphasis on the use of beneficial soil microorganisms present in the rhizospheric region with multifaceted traits which promote plant growth and play a significant role in battling abiotic stress [7,8,9]. Rhizosphere, the layer of soil encasing the plant root, plays an important role in plant growth and development. It is the narrow zone surrounded by plant roots and the hot spot for microorganisms such as bacteria, fungi, nematodes, and algae. It is studied as one of the most complex ecosystems on earth [10,11]. Plant roots exude several metabolites with an abundant supply of carbon such as organic acids, sugars, vitamins, and amino acids which act as signals to attract microbial populations to bolster their proliferation [12,13,14,15]. The total microbial community present in the rhizosphere is called the rhizo-microbiome/rhizosphere microbiome and is divergent from the microbial community of the surrounding soil [16,17].
Within the rhizo-microbiome, a few soil bacteria called plant growth-promoting rhizobacteria (PGPR) colonize the surface of the root system and stimulate the growth and health of the plant by antagonistic and synergistic interactions [7,18,19,20]. Their diversity remains potent with a recurrent shift in community structure and species abundance. These PGPR could be free-living, symbiotic, parasitic, or saprophytic, and play potent roles in promoting plant growth and productivity. Free-living as well as associative and symbiotic rhizobacteria species belonging to the genus Bacillus, Pseudomonas, Azospirillum, Azotobacter, Klebsiella, Enterobacter, Alcaligenes, Arthrobacter, Burkholderia, and Serratia were reported as PGPR [21,22,23].
PGPRs promote plant growth by associative nitrogen fixation, phosphate solubilization, phytohormone production, and volatile organic compounds [24,25,26,27]. PGPR also neutralizes stress in plants created by biotic and abiotic factors by boosting nutrient uptake, osmolyte accumulation, enhanced production of antioxidant enzymes, and metabolites [28,29]. Among several abiotic stresses, salinization of soil increasing continuously and degraded lands all over the globe causes food insecurity by reducing crop productivity [30]. High salt concentration in soils causes osmotic and ionic imbalances, reactive oxygen species (ROS) production, and water stress in plants. This review demonstrates the physiological, biochemical, and molecular mechanisms of salt-tolerant plant growth-promoting rhizobacteria (STPGPR) as emerging biological tools to counterbalance the harmful effects of high salt concentrations [31]. PGPRs play an important role in bioremediation by detoxifying xenobiotics, heavy metals, and pesticides [12,32,33]. PGPRs also revitalize the soil quality by increasing the soil organic content [34].
Numerous literature reviews have discussed the diverse beneficial traits of PGPR and their application as biocontrol agents, but their utilization in agriculture remains challenging worldwide. This may be due to the lack of research on understanding the mechanism of PGPR and plant interactions. The present review will thus attempt to shed more light on the mechanisms demonstrated by PGPR to enhance plant growth and its role in combating various types of abiotic stress to develop strategies for imminent agricultural sustainability. The review article will also delve into the triggers for PGPR colonization, molecular mechanisms, and the impact of PGPR on plant gene expression to elucidate some of the mechanisms by which PGPR enhances plant growth.
- PGPR Mitigating Stress in Plants
2. PGPR Mitigating Stress in Plants
According to Global Agricultural Productivity (GAP), the growth rate of agricultural production must increase by 1.75% annually for there to be enough food to supply the demand of 10 billion people in 2050 [35]. According to Nemecek and Gaillard [36], PGPR greatly influenced farming systems, pedo-climatic conditions, and management techniques. Abiotic factors such as salinity, temperature, drought, fertilizer application, pesticides, heavy metal contamination, and soil pH harm the productivity of crops [6]. Among the abiotic factors, salinization is being considered as the most hazardous stress condition for agricultural productivity [37,38]. Soil salinization has posed a serious threat to food security. It affects the physiological processes, such as aberration in reproductive physiology; the pattern of flowering and fruiting, which affects the crop biomass and yields; and soil processes such as residue decomposition, respiration, denitrification, nitrification, microbial activity, and soil biodiversity [39,40] (Figure 1).
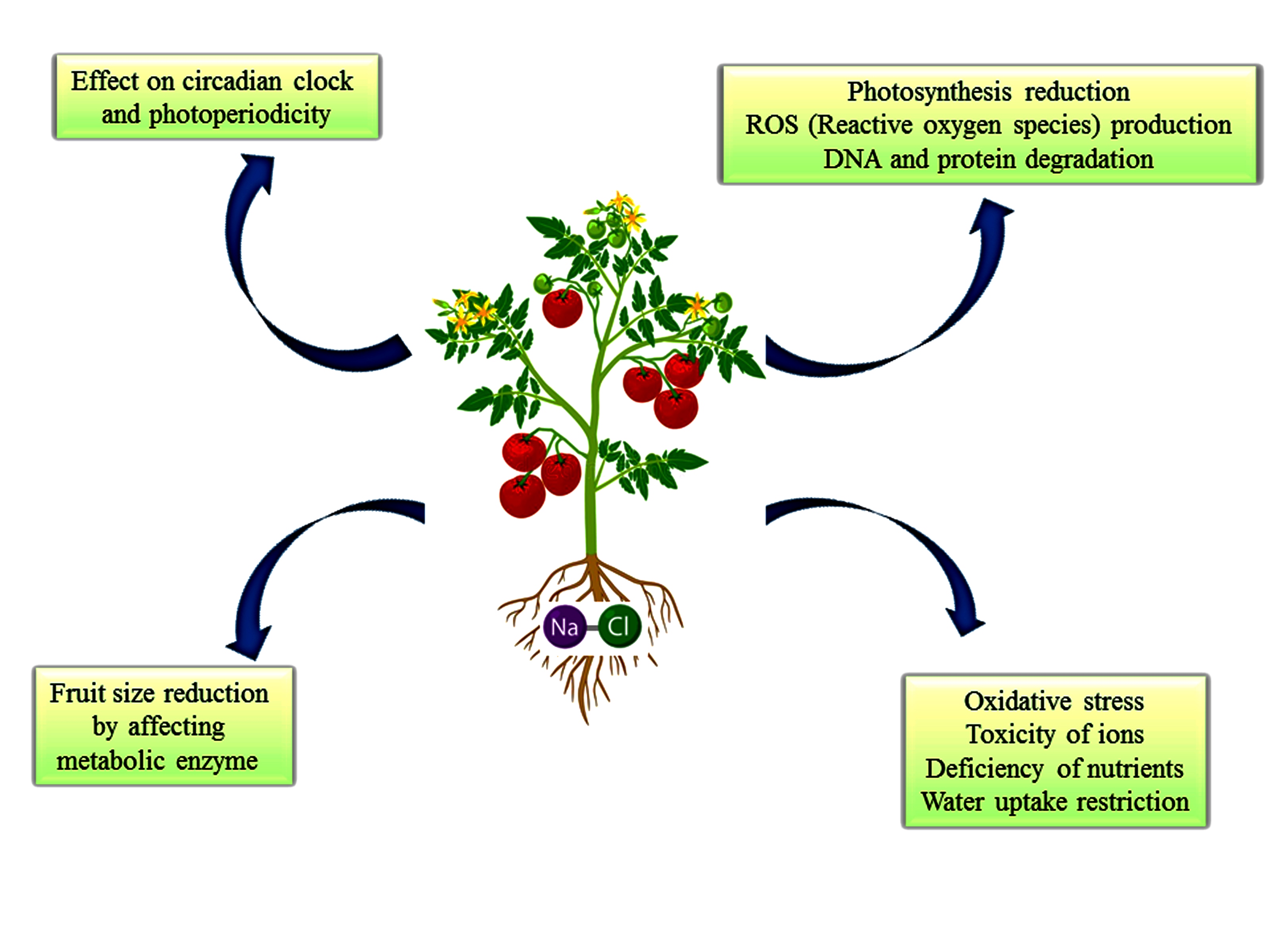

Effect of salinity on plant growth and development.
Fertilizers containing high amounts of salt not only increase the salinity of the soil but also induce osmotic stress in plants, which ultimately hampers plant growth [41,42,43,44]. Reclamation of such saline soils for agricultural activities is time-consuming and not cost-effective [45,46]. The commonly used methods to reclaim saline soils are by using physical (scraping, flushing, and leaching) and chemical (neutralizing agents such as gypsum and lime) processes [47] but these processes possess fewer efficacies in hypersaline soils [48]. Salt tolerant PGPR (ST-PGPR) have been reported to ameliorate salt stress in the plant by direct and indirect mechanisms [30,49,50]. PGPRs produce phytohormones such as cytokinins, auxins, and gibberellins [51], antioxidative enzyme ACC deaminase [50,52], exopolysaccharides [53,54], and osmolytes [55,56] (Figure 2). Pseudomonas, Enterobacter, Bacillus, Klebsiella, Streptomyces, Agrobacterium, and Ochromobacter are reported to improve the productivity of crops under salt stress [57,58]. Rajput et al. [59] reported the enhancement of growth and yield in the wheat crops by an alkaliphilic bacterium Planococcus rifietoensis. ST-PGPR strain Bacillus licheniformis SA03 isolated from saline soil provided increased salt tolerance in Chrysanthemum [60]. A novel salt-tolerant Pseudomonas sp. M30-35 isolated from the rhizosphere of Haloxylon ammodendron reported tolerance capabilities against drought and salt. Bacillus safensis VK isolated from an Indian desert showed salt tolerance capabilities of up to 14% NaCl [61]. Its genome deciphering revealed the presence of several genes which enabled it to function in drought, hypersaline, polyaromatic hydrocarbons (PAHs), and heavy metal contamination.
ST-PGPR Klebsiella sp. IG3 tolerate salinity up to 20% by positively modulating the expression of the WRKY1 (transcription factor dealing with plants reaction to biotic stress) and rbcL (codes for the ribulose-1,5-bisphosphate carboxylase/oxygenase, RuBisCo) genes under saline conditions [62]. A halotolerant PGPR ( Klebsiella sp. D5A) possessed salt tolerance genes and PGP traits such as indole-3-acetic acid (IAA) biosynthesis, phosphate solubilization, acetoin, siderophore production, 2,3-butanediol synthesis, and N2 fixation [63]. Pseudomonas putida and Novosphingobium sp. reduce salt-stress in citrus plants by reducing the level of salicylic acid (SA) and abscisic acid (ABA), the efficiency of photosystem II (Fv/Fm), the accumulation of root chloride and proline, and increasing IAA accumulation under salt stress [64]. Enterobacter sp. UPMR18, a ST-PGPR strain, produces ACC deaminase to improve crop productivity by upregulating ROS pathway genes and enhancing antioxidative enzymes such as APX, SOD, and CAT [65]. The effect of salt stress on plant development is represented in Figure 2.
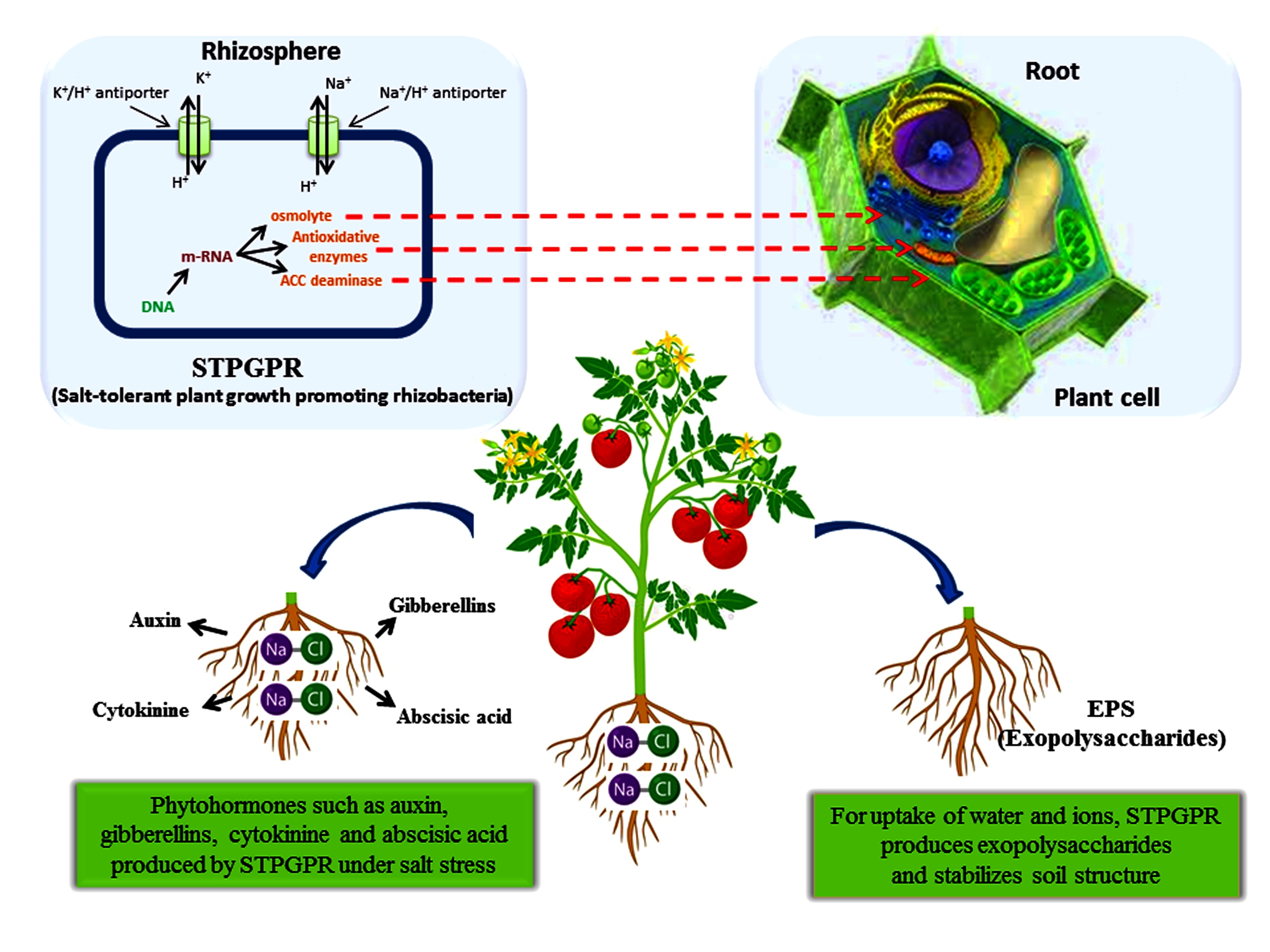

Mitigation of salt stress by STPGPR (salt-tolerant plant growth-promoting rhizobacteria) in plants.
3. PGPR Impact on Plant Gene Expression
- PGPR Impact on Plant Gene Expression
PGPR promotes plant growth promotion by recruiting a variety of direct as well as indirect mechanisms. The most beneficial growth mechanism of PGPR is biological nitrogen fixation, and molecular studies on nitrogen-fixing PGPR isolates revealed the presence of many nif genes coding for nitrogenase enzyme. Apart from nif genes, another gene, fixABCX, was also reported in nitrogen-fixing Rhizobium species and other diazotrophs that coded for a membrane complex, aiding in electron transfer to nitrogenase enzyme [66].
Apart from nitrogen fixation, PGPR isolates are well known for their phosphate solubilization. PGPR solubilizes mineral phosphates by producing gluconic acid catalyzed by the membrane-bound enzyme glucose dehydrogenase and its enzymatic cofactor pyrroloquinoline quinine (PQQ) encoded by pqq operon with six-core genes, namely pqqA, pqqB, pqqC, pqqD, pqqE, and pqqF [67]. Phosphate-solubilization genes, such as gabY, phoC, acpA, napD, and napE genes, and the pqq gene family, were isolated from Pseudomonas cepacia, Morganella morganii, Francisella tularensis, and Burkholderia cepacia [68]. Siderphore production by PGPR is another important characteristic that helps promote plant growth by solubilizing and transporting iron by the formation of soluble Fe
. Siderophores production by PGPR is reported to be due to the up-regulation of sid gene [69]. PGPR alters gene expression in plants by upregulating and downregulating phytohormone genes, metabolism-related genes, stress-response genes, and defense-related genes. Exudates secreted from plants act as signaling molecules and affect the gene expression of microbionts. The root colonization of a halotolerant Rhizobacteria MBE02 on Arachis hypogaea L. (peanut) was reported to reprogram the expression of hormonal signaling genes, which resulted in the overall growth promotion of the peanut. RNA-sequencing analysis revealed the differential expression of 1260 genes in which 979 genes were up-regulated, while 281 were down-regulated by MBE02 treatment. Most of the differentially regulated activated genes were associated with induced systemic resistance (ISR), and hormonal homeostasis in peanut [70]. PGPRs were reported to induce changes in the gene expression of nitrate and ammonium uptake genes in Arabidopsis thaliana [71].
Inoculation of Bacillus amyloliquefaciens SN13 on rice (Oryza sativa) inoculation led to extensive alterations in rice root transcriptome under stress. It induced considerable changes in the expression of a variety of genes involved in photosynthesis, hormone- and stress-response, cell walls, and lipid metabolism under salt stress [72]. PGPR strain Bacillus subtilis JS was reported to up-regulate genes involved in metabolic and cellular processes such as the photosynthetic pathway and photosynthate transport, while it down-regulated the antioxidant enzyme encoding genes such as glutathione S-transferase and methionine-R-sulfoxide reductase [73]. Kerff et al. [74] reported a protein EXLX1 produced by B. subtilis having a structure similar to plant β-expansin which binds to plant cell walls to promote their extension. B. subtilis colonization around A. thaliana plants downregulated the genes related to defense mechanisms in root as well as cell wall-related genes [75,76]. B. subtilis RR4 is reported to suppress various defense-related genes during colonization to roots of rice plantlets to boost plant immunity [77]. Understanding the molecular mechanisms boosting plant growth by PGPR isolates is still evolving and further studies are necessary to verify how PGPR regulates phytobeneficial traits by gene regulation between bacteria and plants during plant colonization.
4. Impact of Environmental Changes on Growth and Development of Microorganism
- Impact of Environmental Changes on Growth and Development of Microorganism
Climatic and soil condition alters the relative abundance and function of soil communities due to differences in their physiology, temperature sensitivity, and growth rates [78,79]. Increments of 5°C in a temperate forest altered the relative abundances of soil bacteria and increased the relation in between the community of bacterial and fungus ratio [80]. Specific microbial groups can regulate ecosystem functions such as N
fixation, nitrification, denitrification, and methanogenesis. Relative changes in the abundance of microorganisms regulate specific processes and show direct impact on the rate of that process. Some processes, such as nitrogen mineralization, are more firmly correlated with abiotic factors, such as moisture and temperature, than the composition of a diverse microbial community [81]. Warming directly alters soil respiration rates of a microbial community due to temperature sensitivity [82]. Clearly, the direct effects of temperature on microbial physiology are mediated by microbial adaptations, their evolution, and specific interactions with time. Changes in temperature and drought are often united with changes in moisture of soil [83]. Less than 30% reduction in the water holding capacity in soil can alter the microbial community, which may shift from one member to another microbial community that remains constant. Microbes continually respond to changes in resources to form complex interaction networks [84,85,86]. Rising temperatures increase carbon allocation symbiotic to parasitic association [87,88] and exacerbate the interaction, negatively or positively, between the plant and its associated community. However, climate conditions, such as soil pH, temperature, and fertility, influence PGPR efficiency and alter the production of biomass, food, and materials from cultivated plants.
5. Conclusion
Among the biological materials used for sustainable agricultural production, PGPR-based bioformulations have sparked immense attention because they provide wide-ranging beneficiary impact on plants using direct and indirect mechanisms they may offer new hope in sustainable agriculture by improving soil fertility, crop productivity, nutrient cycling, and disease tolerance. PGPR also establishes the mutualistic interactions of plant and nutrient absorption such as nitrogen fixation, potassium and phosphorous solubilization, stress tolerance against biotic and abiotic factors, regulation of development and physiology of plants.
References
- Carvalho, F.P. Pesticides, environment and food safety. Food Energy Secur. 2017, 6(2), 48–60, doi:https://doi.org/10.1002/fes3.108
- Chávez-Dulanto, P.C.; Thiry, A.A.; Glorio-Paulet, P.; Vögler, O.; Carvalho, F.P. Increasing the impact of science and technology to provide more people with healthier and safer food. Food Energy Secur. 2021, 10, e259, doi:https://doi.org/10.1002/fes3.259
- IPCC, Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. In: Masson-Delmotte, V.; Zhai, P.; Pirani, A.; Connors, S.L.; Péan, C.; Berger, S.; Caud, N.; Chen, Y.; Goldfarb, L.; Gomis, M.I.; Huang, M.; Leitzell, K.; Lonnoy, E.; Matthews, J.B.R.; Maycock, T.K.; Waterfield, T.; Yelekçi, O.; Yu, R.; Zhou, B., Eds.; Cambridge University Press: 2021.
- Meena, M.; Dubey, M.K.; Swapnil, P.; Zehra, A.; Singh, S.; Kumari, P.; Upadhyay, R.S. The rhizosphere microbial community and methods of its analysis. In Advances in PGPR Research; Singh, H.B.; Sarma, B.K.; Keswani, C., Eds.; CAB International: Wallingford, Oxfordshire, England, 2017a; pp. 275–295, doi:https://doi.org/10.1079/9781786390325.0000
- Khan, N.; Bano, A.; Ali, S.; Md Babar, A. Cross-talk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020a, 90, 189–203, doi:https://doi.org/10.1007/s10725-020-00571-x
- Goswami, M.; Deka, S. Plant growth-promoting rhizobacteria — Alleviators of abiotic stresses in soil: A review. Pedosphere 2020, 30(1), 40–61, doi:https://doi:10.1016/S1002-0160(19)60839-8
- Bhat, M. A.; Kumar, V.; Bhat, M. A.; Wani, I. A.; Dar, F.L.; Farooq, I.; Bhatti, F.; Koser, R.; Rahman, S.; Jan, A.T. Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front Microbiol. 2020, 11, 1952, doi:https://doi.org/10.3389/fmicb.2020.01952
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: recent developments, constraints, and prospects. Sustainability 2021, 13, 1140, doi:https://doi.org/10.3390/su13031140
- Cherif-Silini, H.; Silini, A.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Bouremani, N.; Nowakowska, J.A.; Oszako, T.; Belbahri, L. Tailoring next generation plant growth promoting microorganisms as versatile tools beyond soil desalinization: A road map towards field application. Sustainability 2021, 13(8), 4422, doi:https://doi.org/10.3390/su13084422
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361, doi:https://doi.org/10.1007/s11104-008-9568-6
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37(5), 634–663, doi:https://doi.org/10.1111/1574-6976.12028
- Shukla, K.P.; Sharma, S.; Singh, N.K.; Singh, V.; Tiwari, K.; Singh, S. Nature and role of root exudates: efficacy in bioremediation. J. Biotechnol. 2011, 10(48), 9717–9724, doi:10.5897/AJB10.2552
- Drogue, B.; Combes-Meynet, E.; Moënne-Loccoz, Y.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Control of the cooperation between plant growth-promoting rhizobacteria and crops by rhizosphere signals. In Molecular Microbial Ecology of the Rhizosphere; de Bruijn, F.J., Ed.; John Wiley & Sons: Inc.; NJ, U.S.A., 2013; vol 1-2, pp. 281–294, doi:https://doi: 10.1002/9781118297674
- Meena, M.; Swapnil, P.; Divyanshu, K.; Kumar, S.; Harish, Tripathi, Y.N.; Zehra, A.; Marwal, A.; Upadhyay, R.S. PGPR-mediated induction of systemic resistance and physiochemical alterations in plants against the pathogens: Current perspectives. Basic Microbiol. 2020, 60(10), 828–861, doi:10.1002/jobm.202000370
- Zehra, A.; Raytekar, N.A.; Meena, M.; Swapnil, P. Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: A review. Res. Microb. Sci. 2021, 2, 100054, doi:https://doi.org/10.1016/j.crmicr.2021.100054
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 2013, 8, e55731, doi:https://doi:10.1371/journal.pone.0055731
- Kumar, A.; Dubey, A. Rhizosphere microbiome: Engineering bacterial competitiveness for enhancing crop production. Adv. Res. 2020, 24: 337–352, doi:https://doi.org/10.1016/j.jare.2020.04.014
- Bhardwaj, D.; Ansari, M.W.; Sahoo, R.K.; Tuteja, N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microbial Cell Fact. 2014, 13, 66, doi:https://doi.org/10.1186/1475-2859-13-66
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural uses of plant biostimulants. Plant Soil 2014, 383, 3–41, doi:https://doi.org/10.1007/s11104-014-2131-8
- dos Santos, R.M.; Diaz, P.A.E.; Lobo, L.L.B.; Rigobelo, E.C. Use of plant growth-promoting rhizobacteria in maize and sugarcane: Characteristics and applications. Sustain. Food Syst. 2020, 4, 136, doi:10.3389/fsufs.2020.00136
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28(4), 1327–1350, doi:10.1007/s11274-011-0979-9
- Kumari, P.; Meena, M.; Gupta, P.; Dubey, M.K.; Nath, G.; Upadhyay, R.S. Plant growth promoting rhizobacteria and their biopriming for growth promotion in mung bean (Vigna radiata (L.) R. Wilczek). Agric. Biotechnol. 2018b, 16, 163–171, doi:https://doi.org/10.1016/j.bcab.2018.07.030
- Kumari, P.; Meena, M.; Upadhyay, R.S. Characterization of plant growth promoting rhizobacteria (PGPR) isolated from the rhizosphere of Vigna radiata (mung bean). Agric. Biotechnol. 2018a, 16, 155–162, doi:https://doi.org/10.1016/j.bcab.2018.07.029
- García-Fraile, P.; Menéndez, E.; Rivas, R. Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng. 2015, 2(3), 183–205, doi:10.3934/bioeng.2015.3.183
- Meena, M.; Swapnil, P.; Upadhyay, R.S. Isolation, characterization and toxicological potential of tenuazonic acid, alternariol and alternariol monomethyl ether produced by Alternaria species phytopathogenic on plants. Rep. 2017b, 7, 8777, doi:10.1038/s41598-017-09138-9
- Meena, M.; Swapnil, P.; Zehra, A.; Aamir, M.; Dubey, M.K.; Upadhyay, R.S. Beneficial microbes for disease suppression and plant growth promotion. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Singh, D.; Singh, H.; Prabha, R., Eds.; Springer: Singapore, 2017c; pp. 395–432, doi:https://doi.org/10.1007/978-981-10-6593-4_16
- Chandran, H.; Meena, M.; Sharma, K. Microbial biodiversity and bioremediation assessment through omics approaches. Environ. Chem. 2020a, 1, 570326, doi:10.3389/fenvc.2020.570326
- Kumar, A.; Patel, J.S.; Meena, V.S.; Ramteke, P.W.Plant growth-promoting rhizobacteria: Strategies to improve abiotic stresses under sustainable agriculture. Plant Nutr. 2019, 42(11-12), 1402–1415, doi:10.1080/01904167.2019.1616757
- Kumar, A.; Singh, V.K.; Tripathi, V.; Singh, P.P.; Singh, A.K. Plant growth-promoting rhizobacteria (PGPR): Perspective in agriculture under biotic and abiotic Stress. In New and Future Developments in Microbial Biotechnology and Bioengineering: Crop Improvement Through Microbial Biotechnology, 1st; Prasad, R.; Gill, S.S.; Tuteja, N., Ed.; Elsevier: 2018; pp. 333–342. doi:https://doi.org/10.1016/B978-0-444-63987-5.00016-5
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-tolerant plant growth promoting rhizobacteria for enhancing crop productivity of saline soils. Microbiol. 2019, 10, 2791, doi:https://doi.org/10.3389/fmicb.2019.02791
- Arora, N.K.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P.; Bharti, C. Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. Adv. Res. 2020, 26, 69–82, doi:https://doi.org/10.1016/j.jare.2020.07.003
- Saharan, B.S.; Nehra, V. Plant growth promoting rhizobacteria: A critical review. Life Sci. Med. Res. 2011, 21, 1–30.
- Tak, H.I.; Ahmad, F.; Babalola, O.O. Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. In Reviews of Environmental Contamination and Toxicology; Whitacre, D.M., Ed.; Springer Science Business Media: NewYork, 2013; pp. 33–52, doi:10.1007/978-1- 4614-5577-6_2
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Res. 2018, 206, 131–140, doi:10.1016/j.micres.2017.08.016
- GAP Report. Global Agricultural Productivity Report® (GAP Report®) Global Harvest Initiative, Washington, 2018. Available at: https://globalagriculturalproductivity.org/wp-content/uploads/2019/01/GHI_2018-GAP-Report_FINAL-10.03.pdf
- Nemecek, T.; Gaillard, G. Challenges in assessing the environmental impacts of crop production and horticulture. In Environmental Assessment and Management in the Food Industry; Sonesson, U.; Berlin J.; Ziegler, F., Eds.; Sawston: Woodhead Publishing, 2010; pp. 98–116, doi:10.1533/9780857090225.2.98
- FAO and ITPS. Status of the World’s Soil Resources (SWSR) – Main Report. Rome: Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils. Rome, Italy, 2015; p. 650.
- Shilev, S., Plant-growth-promoting bacteria mitigating soil salinity stress in plants. Applied Sci 2020, 10(20), p.7326.
- Rütting, T.; Aronsson, H.; Delin, S. Efficient use of nitrogen in agriculture. Cycl. Agroecosys. 2018, 110, 1–5, doi:10.1007/s10705-017-9900-8
- Schirawski, J.; Perlin, M.H. Plant microbe interaction 2017 – The good, the bad and the diverse. J. Mol. Sci. 2018, 19(5), 1374, doi:10.3390/ijms19051374
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Plant Biol. 2010, 37(7), 613–620, doi:https://doi.org/10.1071/FP09249
- Herger, G.; Nielsen, R.; Margheim, J. Fertilizer History P3: in WWII Nitrogen Production Issues in Age of Modern Fertilizers, 2015. Available at: http:// cropwatch.unl.edu/fertilizer-history-p3
- Arora, N.K. Impact of climate change on agriculture production and its sustainable solutions. Environmental Sust. 2019, 2, 95–96, doi: 10.1007/s42398-019-00078-w
- Arora, N.K.; Fatima, T.; Mishra, I.; Verma, M.; Mishra, J.; Mishra, V. Environmental sustainability: challenges and viable solutions. Sust. 2018, 1, 309–340, doi:10.1007/s42398-018-00038-w
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Resour. Forum. 2014, 38(4), 282–295, doi:https://doi.org/10.1111/1477-8947.12054
- Kong, X.; Ge, R.; Liu, T.; Xu, S.; Hao, P.; Zhao, X.; Li, Z.; Lei, X.; Duan, H. Super-stable mineralization of cadmium by calcium-aluminum layered double hydroxide and its large-scale application in agriculture soil remediation. Eng. J. 2021, 407, 127178, doi:https://doi.org/10.1016/j.cej.2020.127178
- Ayyam, V.; Palanivel, S.; Chandrakasan, S. Approaches in land degradation management for productivity enhancement. In Coastal Ecosystems of the Tropics – Adaptive Management; Ayyam, V.; Palanivel, S.; Chandrakasan, S., Eds.; Springer Nature: Singapore, 2019; pp. 463–491, doi:https://doi.org/10.1007/978-981-13-8926-9_20
- Rojas-Tapias, D.; Moreno-Galván, A.; Pardo-Díaz, S.; Obando, M.; Rivera, D.; Bonilla, R. Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Soil Ecol. 2012, 61, 264–272, doi:https://doi.org/10.1016/j.apsoil.2012.01.006
- Grover, M.; Ali, S.Z.; Sandhya, V.; Rasul, A.; Venkateswarlu, B. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microbiol. Biotechnol. 2011, 27, 1231–1240, doi:10.1007/s00425-015-2435-9
- Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Al-Huqail, A.A.; Wirth, S.; Egamberdieva, D. The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Microbiol. 2016, 7, 1089, doi:10.3389/fmicb.2016.01089
- Dodd, I.C; Zinovkina, N.Y; Safronova, V.I; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Annals of Applied Biol, 2010,157, pp.361-379, doi.org/10.1111/j.1744-7348.2010.00439.x
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of plant growth by ACC deaminase-producing soil bacteria. In New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research; Bakker, P.A.H.M.; Raaijmakers, J.M.; Bloemberg, G.; Höfte, M.; Lemanceau, P.; Cooke, B.M., Eds.; Springer: Dordrecht, 2007; vol 119, pp. 329–339, doi:https://doi.org/10.1007/978-1-4020-6776-1_8
- Upadhyay, S.K.; Singh, J.S.; Saxena, A.K.; Singh, D.P. Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol, 2012, 14(4), pp.605-611, doi.org/10.1111/j.1438-8677.2011.00533.x
- Timmusk, S.; El-Daim, A.; Moustafa, I.A.; Copolovici, L.; Tanilas, T.; Kännaste, A.; Behers, L.; Nevo, E.; Seisenbaeva, G.; Stenström, E.; Niinemets, Ü. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments. PloS one. 2014, 9, 1-13, org/10.1371/journal.pone.0096086
- Bano, A.; Fatima, M. Salt tolerance in Zea mays (L). following inoculation with Rhizobium and Pseudomonas. Biol Fert Soils, 2009, 45, 405-413, doi.org/10.1007/s00374-008-0344-9
- Upadhyay, S.K.; Singh, D.P. Effect of salt‐tolerant plant growth‐promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol, 2015, 17(1), pp.288-293, doi.org/10.1111/plb.12173
- Sharma, S.; Kulkarni, J.; Jha, B. Halotolerant rhizobacteria promote growth and enhance salinity tolerance in peanut. Microbiol. 2016, 7, 1600, doi:10.3389/fmicb.2016.01600
- Niu, X.; Song, L.; Xiao, Y.; Ge, W. Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Microbiol. 2018, 8, 2580, doi:10.3389/fmicb.2017.02580
- Rajput, L.U.B.N.A.; Imran, A.; Mubeen, F.; Hafeez, F.Y. Salt-tolerant PGPR strain Planococcus rifietoensis promotes the growth and yield of wheat (Triticum aestivum) cultivated in saline soil. Pak. J. Bot. 2013, 45, 1955–1962.
- Zhou, C.; Zhu, L.; Ma, Z.; Wang, J. Bacillus amyloliquefaciens SAY09 increases cadmium resistance in plants by activation of auxin-mediated signaling pathways. Genes 2017a, 8(7), 173, doi:https://doi.org/10.3390/genes8070173
- Kothari, V.V.; Kothari, R.K.; Kothari, C.R.; Bhatt, V.D.; Nathani, N.M.; Koringa, P.G.; Joshi, C.G.; Vyas, B.R. Genome sequence of salt-tolerant Bacillus safensis strain VK, isolated from Saline Desert Area of Gujarat, India. Genome Announc. 2013, 1(5), e00671-13, doi:10.1128/genomeA.00671-13
- Sapre, S.; Gontia-Mishra, I.; Tiwari, S. Klebsiella confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbiol. Res. 2018, 206, 25–32, doi:10.1016/j.micres.2017.09.009
- Liu, W.; Wang, Q.; Hou, J.; Tu, C.; Luo, Y.; Christie, P. Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella D5A. Sci. Rep. 2016, 6, 26710, doi:10.1038/srep26710
- Vives-Peris, V.; Gomez-Cadenas, A.; Perez-Clemente, R.M. Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria Pseudomonas putida and Novosphingobium Plant Cell Rep. 2018, 37, 1557–1569, doi:10.1007/s00299-018-2328-z
- Habib, S.H.; Kausar, H.; Halimi, M. Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Res. Int. 2016, 2016, 6284547. doi: 10.1155/2016/6284547
- Mahmud, K.; Makaju, S.; Ibrahim, R.; Missaoui, A. Current progress in nitrogen fixing plants and microbiome research. Plants (Basel, Switzerland) 2020, 9(1), 97, doi:https://doi.org/10.3390/plants9010097
- Matsushita, K.; Shinagawa, E.; Ameyama, M. D-Gluconate dehydrogenase from bacteria, 2-keto-d-gluconate-yielding, membrane-bound. Methods Enzymol. 1982, 89, 187–193, doi:https://doi.org/10.1016/S0076-6879(82)89033-2
- Alaylar, B.; Egamberdieva, D.; Gulluce, M.; Karadayi, M.; Arora, N.K. Integration of molecular tools in microbial phosphate solubilization research in agriculture perspective. World J Microbiol Biotechnol,2020,36,1-12, doi.org/10.1007/s11274-020-02870-x
- Ovaa, W.; Bitter, W.; Weisbeek, P.; Koster, M. Multiple outer membrane receptors for uptake of ferric pseudobactins in Pseudomonas putida Mol. Gen. Genet. 1995, 248, 735–743, doi:https://doi.org/10.1007/BF02191714
- Sharma, S.; Chen, C.; Navathe, S. Chand, R.; Pandey, S.P. A halotolerant growth promoting rhizobacteria triggers induced systemic resistance in plants and defends against fungal infection. Rep. 2019, 9, 4054, doi:https://doi.org/10.1038/s41598-019-40930-x
- Calvo, P.; Zebelo, S.; McNear, D.; Kloepper, J.; Fadamiro, F.Plant growth-promoting rhizobacteria induce changes in Arabidopsis thaliana gene expression of nitrate and ammonium uptake genes. Plant Interact. 2019, 14(1), 224–231, doi:10.1080/17429145.2019.1602887
- Chauhan, P.S.; Lata, C.; Tiwari, S.; Chauhan, A.S.; Mishra, S.K.; Agrawal, L.; Chakrabarty, D.; Nautiyal, C.S. Transcriptional alterations reveal Bacillus amyloliquefaciens-rice cooperation under salt stress. Rep. 2019, 9(1), 11912, doi:https://doi.org/10.1038/s41598-019-48309-8
- Kim, J.S.; Lee, J.; Seo, SG. Lee, C.; Woo, S.Y.; Kim, S.H. Gene expression profile affected by volatiles of new plant growth promoting rhizobacteria, Bacillus subtilis strain JS, in tobacco. Genes Genom. 2015, 37, 387–397, doi:https://doi.org/10.1007/s13258-015-0267-4
- Kerff, F.; Amoroso, A.; Herman, R.; Sauvage, E.; Petrella, S.; Filée, P.; Charlier, P.; Joris, B.; Tabuchi, A.; Nikolaidis, N.; Cosgrove, D.J.Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization. Natl. Acad. Sci. U.S.A. 2008, 105, 16876–16881, doi:https://doi.org/10.1073/pnas.0809382105
- Lakshmanan, V.; Bais, H.P.Factors other than root secreted malic acid that contributes toward Bacillus subtilis FB17 colonization on Arabidopsis Plant Signal. Behav. 2013, 8, 11, doi:10.4161/psb.27277
- Blake, C.; Christensen, M.N.; Kovács, A.T. Molecular aspects of plant growth promotion and protection by Bacillus subtilis. Plant Microbe Interact. 2021, 34(1), 15–25, doi:https://doi.org/10.1094/MPMI-08-20-0225-CR
- Rekha, K.; Kumar, R.M.; Ilango, K.; Rex, A.; Usha, B.Transcriptome profiling of rice roots in early response to Bacillus subtilis (RR4) colonization. Botany 2018, 96, 749– doi:https://doi.org/10.1139/cjb-2018-0052
- Whitaker, J.; Ostle, N.; Nottingham, A.T.; Ccahuana, A.; Salinas, N.; Bardgett, R.D.; Meir, P.; McNamara, N.P. Microbial community composition explains soil respiration responses to changing carbon inputs along an Andes‐to‐Amazon elevation gradient. Ecol. 2014, 102(4), 1058–1071, doi:https://doi.org/10.1111/1365-2745.12247
- Chen, M.; Cao, H.; Peng, H.; Hu, H.; Wang, W.; Zhang, X. Reaction kinetics for the biocatalytic conversion of phenazine-1-carboxylic acid to 2-hydroxyphenazine. PLoS One 2014, 9(6), e98537, doi:https://doi.org/10.1371/journal.pone.0098537
- DeAngelis, K.M.; Pold, G.; Topçuoğlu, B.D.; van Diepen, L.T.; Varney, R.M.; Blanchard, J.L.; Melillo, J.; Frey, S.D. Long-term forest soil warming alters microbial communities in temperate forest soils. Microbiol. 2015, 6, 104, doi:https://doi.org/10.3389/fmicb.2015.00104
- Hooper, D.U.; Chapin Iii, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; Schmid, B. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Monogr. 2005, 75(1), 3–35, doi:https://doi.org/10.1890/04-0922
- Hagerty, S.B.; Van Groenigen, K.J.; Allison, S.D.; Hungate, B.A.; Schwartz, E.; Koch, G.W.; Kolka, R.K.; Dijkstra, P. Accelerated microbial turnover but constant growth efficiency with warming in soil. Clim. Chang. 2014, 4(10), 903–906, doi:https://doi.org/10.1038/nclimate2361
- Briones, M.J.I.; McNamara, N.P.; Poskitt, J.; Crow, S.E.; Ostle, N.J. Interactive biotic and abiotic regulators of soil carbon cycling: evidence from controlled climate experiments on peatland and boreal soils. Change Biol. 2014, 20(9), 2971–2982, doi:https://doi.org/10.1111/gcb.12585
- Nuccio, E.E.; Hodge, A.; Pett‐Ridge, J.; Herman, D.J.; Weber, P.K; Firestone, M.K. An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Microbiol. 2013, 15(6), 1870–1881, doi: https://doi.org/10.3389/fmicb.2017.01120
- Clemmensen, K.E.; Bahr, A.; Ovaskainen, O.; Dahlberg, A.; Ekblad, A.; Wallander, H.; Stenlid, J.; Finlay, R.D.; Wardle, D.A; Lindahl, B.D. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 2013, 339(6127), 1615–1618, doi:10.1126/science.1231923
- Moore, J.A.M.; Jiang, J.; Post, W.M; Classen, A.T. Decomposition by ectomycorrhizal fungi alters soil carbon storage in a simulation model. Ecosphere 2015, 6(3), 1–16, doi: https://doi.org/10.1890/ES14-00301.
- Hawkes, C.V.; Hartley, I.P.; Ineson, P.; Fitter, A.H. Soil temperature affects carbon allocation within arbuscular mycorrhizal networks and carbon transport from plant to fungus. Chang. Biol. 2008, 14(5), 1181–1190, doi:https://doi.org/10.1111/j.1365-2486.2007.01535.x
- Nazir, R.; Warmink, J.A.; Boersma, H.; Van Elsas, J.D. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 2009, 71(2), 169–185, doi:1111/j.1574-6941.2009.00807.x
