Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Chien-Yen Chen and Version 2 by Rita Xu.
Worldwide biodiversity loss points to a necessity of upgrading to a fast and effective monitoring method that can provide quick conservation action. Newly developed environmental DNA (eDNA) based method found to be more cost-effective, non-invasive, quick, and accurate than traditional monitoring (spot identification, camera trapping).
- environmental DNA application
- biodiversity monitoring
- invasive species
1. Introduction
The loss of biodiversity has been one of the most serious concerns worldwide. The world has been losing its biodiversity due to a target to fulfilling high demands of satisfaction by the human race which in turn is incurring expensive and detrimental demands to nature [1]. According to IPBES (The Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services) report, 25% of animals and plants are already threatened with extinction [2]. The wild animals and plants, as well as domestic ones, are facing a fight for survival due to anthropogenic activity. In the next thirty years, 30–50 % of plant species will become extinct [3]. The current rate of extinction is 1000 to 10,000 times greater than the natural extinction rate on our planet [2]. This is an extremely serious issue that will be more severe in the coming days. However, to mitigate this issue, we need to initiate a monitoring program at both local and global levels. In designing such a monitoring system, we need to consider the development of a fit-for-purpose, accurate and cost-effective technique for the detection of species, assessment of biodiversity and study of species interactions.
Environmental DNA, known as eDNA, is shed by organisms during their existence in nature [4]. During their lifespan, organism shed DNA wherever they have been present for a moments. The collection and analysis of these environmental samples and monitoring of the ecosystem without harming organisms is the basis of eDNA study. Recently eDNA, provided valuable contribution to both aquatic and terrestrial monitoring [5][6][5,6]. Originally, the eDNA-based species detection was a microbiological study, dating back in 1987 [7] and the use of eDNA to detect macro-organism directly from water sample came to the front in early 2008 with detection of aquatic invasive species [8]. Later on, the methodology was updated and reinforced by some pioneer studies [9][10][9,10]. Afterward, rate of eDNA release, degradation, persistence as well as the changes in concentration with organism abundance were explored [11][12][13][14][15][16][11,12,13,14,15,16]. However, as more studies incorporated the use of eDNA approaches, terminology quickly diverged, becoming more convoluted or generally misunderstood [17][18][17,18]. What quickly followed was two distinct schools of thought: (i) those who view eDNA as relating to any DNA originating from environmental samples (eDNA sensu lato: [4]), and (ii) research referring to eDNA originating from macro-organisms specifically (eDNA sensu stricto). Researchers use eDNA for species detection to reveal many critical ecological questions, such as studies of population genetics, abundance and habitat preference, detection of unrecorded populations, understanding behavioral biology, monitoring of reproductive migration, pathogens, terrestrial plant community, biodiversity of marine and river ecosystem, nutrient quality, assessment of coral ecosystem, etc. [19]. Moreover, the novel eDNA-based approaches have been used to solve some critical conservation issues such as the detection of rare and endangered species [20], invasive species [21], monitoring whole biodiversity [22][23][22,23], study of anthropogenic effect [24], ecosystem health [25] and disease [26]. As eDNA-based methods are emerging rapidly as a multidisciplinary branch of science (Figure 1), it is necessary to evaluate the recent advancements for proper implementation.
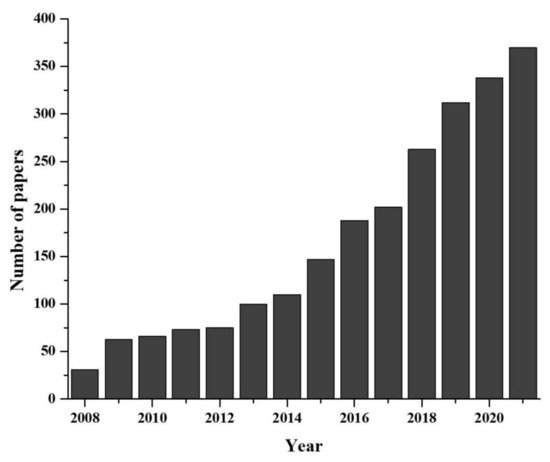
Figure 1. Developmental progress of eDNA technique in last two decades (data collected from PUBMED advanced search with “environmental DNA or eDNA” as title).
2. Overview of Sampling and Laboratory Protocol for eDNA
The detection of biological signature from eDNA traces has been reported from different environment (e.g., water, soil, air, snow, and even in drinking water) [27], where the sampling approaches and extracting protocols of eDNA have indicated ‘required modifications’ depending on sample type and interest [16].
2.1. Collection and Accumulation of eDNA Samples
The long-time exposure and abundance of target organisms strongly increase the amount of eDNA into the environment [28], where detection probability was reported to be higher near the habitat [29]. Nevertheless, organisms in low abundance can be traced with meticulous experimental design [30]. The occurrence of the targeted eDNA in the environment depends upon their life history, body size, behavior, seasonal and reproductive activity (e.g., eDNA amount observed is higher during the breeding season) [6]. The persistence may depend upon the physicochemical factors of the environment (e.g., temperature, pH, and oxygen) [29]. Not only the physicochemical characteristics of the medium, but also the mobilization of the medium (e.g., intra-medium: water to water; inter-medium: soil to water) influences the existence/persistence of eDNA in the environment, including false-positive detection due to multiple factors (e.g., medium current, settlement, predator, anthropogenic activity, etc.) [28].
2.1.1. Aquatic Environment
Generally, in aquatic biomes, the existence of eDNA differs according to sampling zones (e.g., littoral, limnetic, and intertidal) [6][16]. The different effective samplers (e.g., Nansen metal water sampler, Bucket and van Dorn sampler, Kemmerer type water sampler, Niskin water sampler, Bottles, PVC pole, and Polyethylene Nalgene bottles, etc.) are used in different studies for eDNA sampling [31][32]. In the case of lentic ecosystems, eDNA deposits in sediments, since water is stagnant, whereas long-dated eDNA is observed in significantly higher amounts in sediment zone compared to photic zone [33]. Here, a well cleaned DNA free bottle or one-time use sampler is suitable for the collection, whereas a sampler equipped with pole/rope-like structure is used for benthic water sampling [34]. On the other hand, in the lotic ecosystem, a filter funnel can be used against the flow to collect eDNA from water; however, the chances of false-positive results (due to transportation of eDNA) should be kept in consideration.
The sample can be processed through filtration/centrifugation/ultra-centrifugation/ precipitation steps after collection (if the accumulation step is not performed) [32]. The filtration technique (as it processes larger volumes of water at once) is the most common method adopted for accumulation of DNA into filter paper (0.2–3.0 µm size) [32][35]. The main problem of this technique is clogging, in particular for smaller pore filters; here the use of two or more separate filter papers for samples can be adopted. Moreover, conventional filtration, enclosed sterivex filters (pressure mediated filter without electricity) are highly effective and advanced techniques in remote field surveys [35]. The ethanol precipitation, centrifugation, and ultra-centrifugation steps are suitable for places where long-time access is difficult and when the targeted DNA is present in high concentration (because those generally process less volume of sample) [32][35]. In the precipitation step, ethanol or isopropanol are required to accumulate DNA; however, the centrifugation and ultra-centrifugation steps require no chemicals [16]. Furthermore, the detection probability is correlated with sample volume, although it is necessary to optimize it depending on target organism [36]. Tsuji et al. [32] reviewed the protocols on eDNA studies in water, which indicated that over 78% of cases used the filtration method, followed by ethanol precipitation (13%) and centrifugation (4%). The most common type of filter paper is used as cellulose nitrate (CN) 0.45 µm pore size, although other types (0.45 µm mixed cellulose ester membrane, 0.7 µm glass microfiber, etc.) are also considered [32][37].
2.1.2. Terrestrial Environment
The selection of different soil layers eventually depends on targeted taxa. The soil may be collected using a sterile digger or debris metal screens (to remove large particles), and collected soil needs to be kept in a dark box (containing ice) for the transpiration purpose to the laboratory as soon as possible for DNA extraction [38]. In another technique, the soil sample can be dissolved in water by agitation, followed by filtration (like water) to concentrate the DNA into filter paper. The sterile tubes, modified plastic syringes, and drilling core samples can be inserted into the sediment to withdraw the sediment samples for eDNA study. In the case of air sampling, especially designed volumetric samplers can be used [23].
2.1.3. Extraction of eDNA from Other Organisms without Isolating Target Taxa
The eDNA also can be extracted from a non-target organism to study species interaction data of target organisms, such as feces to study the dietary information, insect-derived DNA (iDNA) to study mammalian diversity, flower to study plant-pollinators-interaction, water from sponges to study marine diversity, etc. [39][40][41][42].
2.2. Preservation of Samples in eDNA Technique
The quality and quantity of DNA undergoes degradation/change due to microbial activity, mechanical forces, chemical reactions, etc., hence the preservation of sample is an essential step for experimental design [37]. On-site, if filtration and/or preservation is not possible then samples should be stored in dry-ice or in a styrofoam box with cooling elements (just after collection) and at −20 °C in the laboratory (not more than 8 h) [32][37]. Dried or semi-dried samples (e.g., soil, feces) also can be stored in a pouch containing silica beads. The filter is generally preserved by freezing in a liquid medium or dry medium. The liquid preservative (e.g., ethanol, Longmire’s buffers, cetyl trimethyl ammonium bromide (CTAB), ATL lysis buffer (Qiagen, Hilden, Germany), etc.) are effective to store DNA present on the filter paper [32][37]. Alternatively, filter paper (wrapped in aluminum foil or directly) can be placed in a silica gel containing packet (to keep it moisture free) [37].
2.3. The eDNA Analysis in Laboratory
Samples should be stored in dry preservative, which later on can undergo extraction process directly (e.g., soil sample) [43], whereas the filter paper should be stored in ethanol or other liquid media, before DNA extraction, and should be kept in an open micro-centrifuge tube under a fume hood to let the liquid evaporate properly. On the other hand, for centrifugation, ultra-centrifugation, and precipitation steps, DNA can be extracted from pellets [37]. It is important to mention that both conventional (e.g., CTAB) and commercial kits (time-saving and less hazardous) are available (e.g., DNeasy blood tissue kit, Qiagen) for DNA extraction [32][37].
In the case of single species detection, eDNA is subjected to amplification with species-specific primer [44]. Here, conventional PCR (cPCR) can be used to conduct a ‘presence and absence’ study whereas quantitative PCR (qPCR) is more preferred for quantification of targeted DNA and elimination of false positive or negative results [16][45]. In qPCR, probes are one of the best option to identify particular species, although intercalating dye (e.g., SYBR Green) can be used instead of probes for cost-effectiveness [45]. However, the droplet digital PCR (ddPCR) (sensitive PCR) has better species specificity and quantification accuracy than the formers [31]. Recently, CRISPR-Cas is gaining popularity in eDNA-based species detection [46]. Moreover, in all cases, positive and negative controls should be maintained [44].
The evolution of technology has allowed to introduce a new high-throughput sequencing (HTS) platform enabling analysis and identification of whole communities, commonly termed DNA metabarcoding [4]. The HTS platform can produce billions of sequences in a single run, allowing analyzing several samples in parallel and identifying several species in each sample. Such an advancement leads to an increase in the computational load, and it is imperative to move toward high-performance computing. Furthermore, DNA metabarcoding is a widely tested and validated approach for processing mixed taxon. The community detection through DNA metabarcoding relies on “universal” primers (i.e., non-specific primers), therefore introducing amplification bias when the primers match some taxa better than others during PCR amplification [38]. This bias might be counteracted using hybridization probes by allowing the targeted capture of barcoding genes. An alternative is to sequence directly the extracted bulk DNA without PCR [47]. These metagenomic techniques overlook most of the problems associated with PCR-based metabarcoding, such as the loss of some taxonomic groups due to primer binding sites [48], and they are well established for bacterial communities and recently applied to eukaryotes [38]. An advisable procedure is to design a metabarcoding-based study in order to be ecosystem-specific and target-gene primer sets. Considering the desired ecosystem and taxonomic context, in-silico and in-vitro tests should be performed to validate the applicability of the primer pairs. PCR primers (species-specific or universal) can be designed manually or using software (e.g., Primer Express 3.0, 3.0.1, NCBI primer blast, allele ID) [48]. Furthermore, practitioners can also consult for standard manual or technical advice to eDNA societies/private sectors, such as DNAqua-Net (https://dnaqua.net/), EnviroDNA (https://www.envirodna.com/), CaleDNA (https://ucedna.com/), The eDNA society, Japan (https://ednasociety.org/en/), etc., and all of the links are accessed on 20 November 2021. Moreover, a general outline of eDNA based technique is presented in Figure 2.
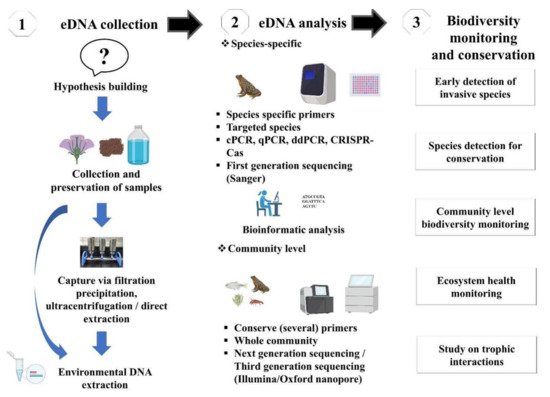

Figure 2. Schematic workflow of eDNA-based studies and its application in biodiversity monitoring and conservation.
3. Precautions in eDNA Study
Contamination can arise from various sources (indicated below) at any stage during the sampling and laboratory analysis which can distort the result precision.
3.1. Precautions in Field
In the field, contamination is a major issue for inter and intra sampling. The sterilized sampling boots or sterile chest waders should be used when the researcher is required to reach deep into the sampling site for sample collection [44]. It is highly recommended that all tools should be sterilized with 10–50% bleach solution followed by deionized water (>2 times) [30]. Special care should be taken during opening the filter paper (sterile filters should be preferred) from the package and after filtration, as filter paper should be removed using previously sterilized forceps.
3.2. Precautions in Laboratory
In the laboratory, although a specific or universal primer is present, there is a chance of false-positive or false-negative detection. Types of errors in DNA-based detection are well-reviewed by Darling et al. [49]. To obtain accurate results in the laboratory, there are several precautions that should be taken under consideration: (i) cleanliness, (ii) wearing of clean clothes, (iii) one-time use of gloves and a facial masks, (iv) cleaning of the surface of workplace using chemical (e.g., DNA Away, Decon 90, DNA-exitusPlus and bleach solution) and physical methods (UV light), (v) DNA extraction in a contamination-free zone (vi) restriction of movement during while handling PCR, (vii) use of proper control samples, and (viii) totally unidirectional workflow, [30][44].
