Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Chao Wu and Version 2 by Catherine Yang.
ATP-binding cassette (ABC) transporters, a large class of transmembrane proteins, are widely found in organisms and play an important role in the transport of xenobiotics. Insect ABC transporters are involved in insecticide detoxification and Bacillus thuringiensis (Bt) toxin perforation. The complete ABC transporter is composed of two hydrophobic transmembrane domains (TMDs) and two nucleotide binding domains (NBDs).
- insect
- ABC transporters
- structure
- Bt Insecticidal Activity
1. Introduction
ATP-binding cassette (ABC) proteins comprise an extensive and variable transporter superfamily within P-loop motif and are found in all living organisms [1][2][3][1,2,3]. Studies on ABC transporters began in the early 1970s with the biochemical characterization of substrate-binding protein-dependent transport in Escherichia coli that was directly energized by hydrolysis of ATP [4][5][4,5]. In 1982, cytoplasmic membrane-associated transporter genes in the histidine transport system of Salmonella typhimurium (coded by the hisP gene) and maltose-maltodextrin transport system of E. coli (coded by the malK gene) were cloned [6][7][6,7]. Concurrently, in mammalian cells, the gene encoding permeability, glycoprotein (P-gp, a large glycosylated membrane protein related to multi-drug resistance) was identified and cloned in 1985 [8][9][8,9]. Eventually, substrate-binding transport proteins with ATP-binding subunits were found to constitute a large superfamily of transport proteins and termed ABC transporters in 1990 [10]. On the basis of differences in the ATP-binding sites among insect ABC transporters, the superfamily can be divided into eight subfamilies (ABCA to ABCH) [11].
2. Structure and Mechanism of ABC Transporters
Structural models of ABC transporters are based on the crystal structure of different bacterial proteins that act as importers such as vitamin B12 transporter BtuCDF from E. coli and exporters such as the multidrug exporter Sav1866 from Staphylococcus aureus or related flippases such as MsbA lipid flippase from E. coli [12][13][14][29,30,31]. On the basis of their architecture and biochemical activity, the ABC importers have been divided into type I and type II [15][16][17][12,32,33]. The energy coupling factor (ECF) transporters, which differ structurally and functionally from other ABC importers, are sometimes considered as type III ABC importers [18][19][20][21][34,35,36,37]. However, ABC importers have only been confirmed in prokaryotes, not in eukaryotes [11][22][11,15]; therefore, in this review, we focus only on ABC exporters.
The structure of ABC transporters is highly conserved among most eukaryotic organisms, including insects. A functional ABC transporter is characterized by the presence of a P-type traffic ATPase, which comprises two cytosolic nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs) [1][15][23][1,12,38] (Figure 1A). The four domains of a functional transporter (2TMDs-2NBDs) are combined in a single polypeptide, forming a full transporter (FT), whereas a half transporter (HT) contains one TMD and one NBD, which are sometimes encoded as separate polypeptides and then fused into multidomain proteins. For ATP binding and hydrolysis, the HT must become a functional transporter by forming homo- or heterodimeric complexes. The NBD contains several highly conserved nucleotide-binding sequences such as the Walker A and B motifs, common in nucleotide-binding proteins (the Walker B motif also provides the catalytic base); d-loop, which contains an aspartate residue and is responsible for forming a salt bridge; Q-loop, which contains a glutamate residue and acts as the attacking nucleophile in ATP hydrolysis; H motif, which has an invariant histidine active site that may be involved in maintaining the stability of the pre-hydrolytic state; and an α-helical region that has the ABC signature sequence (LSGGQ motif) [15][24][25][12,13,39]. The ABC exporter fold, a prominent quaternary structure in TMDs in all ABC exporters, is characterized by 12 transmembrane helices and acts as a switch between different conformational changes and initiates substrate translocation [15][12].
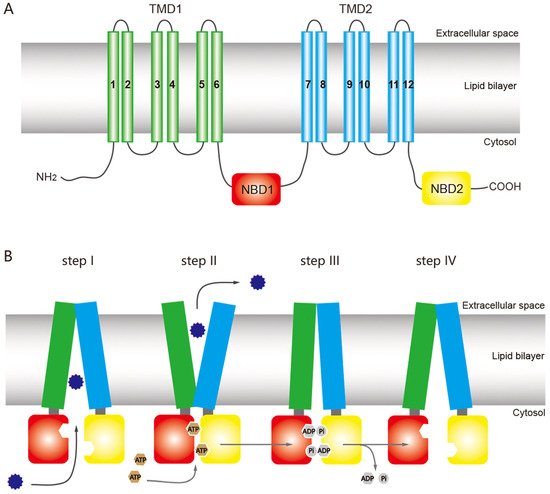

Figure 1. General structure of an ATP-binding cassette (ABC) full transporter (ABC exporter) and the ATP-switch model for the transport mechanism of ABC transporters. (A) Typical ABC full transporter with two transmembrane domains (TMDs), TMD1 (green) and TMD2 (sky blue), and two nucleotide-binding domains (NBDs), NBD1 (red) and NBD2 (yellow). Each transmembrane domain (TMD) contains six transmembrane helices. The “long” multidrug-resistance associated proteins (MRPs) of the ABCC subfamily contains an additional TMD (TMD0) at the N terminus [26][46]. (B) The ATP-switch model [27][14] includes (I) binding of the substrates (12-point blue circle) to the TMDs; (II) subsequent structural changes to the NBDs (red and yellow), hydrolysis of ATP (brown circles), followed by closed dimer formation of the NBDs and major conformational change in the TMDs, which initiates substrate translocation; (III) the ATP is hydrolyzed (gray circles), releasing ADP and Pi, and (IV) finally destabilization of the closed dimer restores its initial open dimer configuration for another new cycle. This figure is drawn by following the previous report of ABC transporter by Dermauw & Van Leeuwen [27][14].
ABC transporters have a common mechanism for exporting substrates across the membrane by hydrolyzing ATP as a pump, but other models have been proposed for the ABC transporter mechanism based on structural and biochemical evidence, including the ATP-switch [25][39], alternating site [28][40], constant contact [29][30][41,42], and thermodynamic models [31][43]. Among these models, the ATP-switch model provides a reasonable framework for the transport mechanism [25][32][33][39,44,45] in which repeated communication between NBDs and TMDs occurs in both directions and involves only non-covalent conformational changes. The transport process is initiated by the binding of the substrate to the TMDs, and subsequent structural changes are transmitted to the NBDs, which include ATP-binding and closed dimer formation of the NBDs. Then the closed NBD dimer induces a substantial conformational change in the TMDs. This conformational change initiates translocation of the substrate through a rotation of the TMDs and opening toward the extracellular milieu. Finally, the ATP is hydrolyzed, releasing ADP and Pi and destabilizing the closed dimer conformation to restore its open dimer configuration for another new cycle [25][32][33][39,44,45] (Figure 1B).
3. ABC Transporter Subfamilies in Insects
In recent years, with the large-scale development of genome sequencing technology, the sequencing results have shown that ABC transporter genes are highly conserved in many insects (Figure 2). Aside from some important discoveries on the function of some ABC transporters in insects, however, knowledge on the role and function of these proteins is still limited. ABC transporters of numerous important agricultural pests and model insects, such as D. melanogaster, B. mori, Helicoverpa armigera and Plutella xylostella, have been reported (Table 1).
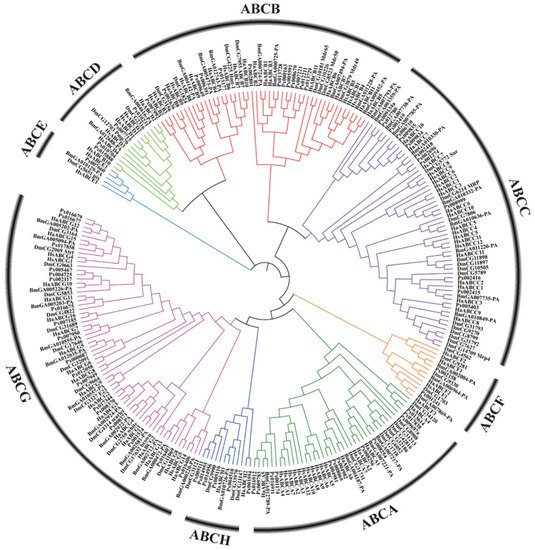

Figure 2. Phylogenetic tree based on amino acid sequences of 262 ABC transporters (Supplementary Material 1) from several insects and humans. The sequences were aligned using MUSCLE. The evolutionary history was inferred using the neighbor-joining method and MEGA-X with 1000 bootstrap replicates. All positions with less than 95% site coverage were eliminated. Species codes: Ha, Helicoverpa armigera; Bm, Bombyx mori; Px, Plutella xylostella; Hs, Homo sapiens; Dm, Drosophila melanogaster.
Table 1.
Distribution of genes among ABC transporter subfamilies for different arthropods and
Homo sapiens
.
| Organisms | A | B | C | D | E | F | G | H | Total | References |
|---|
| Homo sapiens | 12 | 11 | 12 | 4 | 1 | 3 | 5 | 0 | 48 | [1] | ||
| Drosophila melanogaster | 10 | 8 | 14 | 2 | 1 | 3 | 15 | 3 | 56 | [1] | ||
| Anopheles gambiae | 9 | 5 | 13 | 2 | 1 | 3 | 16 | 3 | 52 | [34] | [47] | |
| Daphnia pulex | 4 | 7 | 7 | 3 | 1 | 4 | 24 | 15 | 65 | [35] | [48] | |
| Pediculus humanus humanus | 2 | 6 | 5 | 2 | 1 | 3 | 13 | 6 | 40 | a | [36] | [49] |
| Apis melifera | 3 | 5 | 9 | 2 | 1 | 3 | 15 | 3 | 41 | [37] | [50] | |
| Bombyx mori | 7 | 9 | 11 | 2 | 1 | 3 | 16 | 2 | 51 | b | [37][38][39] | [50,51,52] |
| Tribolium castaneum | 10 | 6 | 35 | 2 | 1 | 3 | 13 | 3 | 73 | [40] | [20] | |
| Tetranychus urticae | 9 | 4 | 39 | 2 | 1 | 3 | 23 | 22 | 103 | [41] | [53] | |
| Chrysomela populi | 5 | |||||||||||
| Helicoverpa zea | ||||||||||||
| 7 | ||||||||||||
| 11 | ||||||||||||
| 11 | ||||||||||||
| 2 | ||||||||||||
| 1 | 3 | 17 | 2 | 54 | [ | 39 | ] | [ | 52 | ] | ||
| Acyrthosiphon pisum | 11 | 9 | 16 | 2 | 1 | 4 | 19 | 9 | 71 | [51][52] | [63,64] |
a Includes two uncharacterized ABC transporters. b The number of B. mori ABC transporter genes in the different subfamilies is mainly based on the latest report [39][52], but were reported previously [37][50].
4. ABC Transporter Roles in Bt Insecticidal Activity
The ABC transporters that act as Bt-toxin receptor belong to subfamilies A, B, C and G (Table 2). With the long-term cultivation of Bt cotton, field resistance to Cry2Ab toxin evolved in H. armigera. The resistance of two H. armigera strains to Cry2Ab was an independent evolutionary event involving different deletion mutations, which were located in different exons of the same ABCA2 gene [53][68]. A homologous deletion in HaABCA2 was also detected in resistant lines of H. punctigera Cry2Ab. A similar situation was also found in resistant lines of Pectinophora gossypiella; a loss of PgABCA2 exon 6 was caused by alternative splicing in resistant larvae of laboratory-selected strains in Arizona and field-selected strains in India [54][118]. In addition, insect ABCA2 was expressed in the midgut where the Cry2Ab toxin binds [53][55][68,119]. A bioassay of two strains of H. armigera with CRISPR/cas9 knockout of HaABCA2 showed that HaABCA2 plays an important role in mediating resistance to Cry2Aa and Cry2Ab [56][120].
Table 2.
Bt toxins and their possible corresponding ABC transporters as receptors in insects.
| Bt toxin | Receptor | Target pest | Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cry1Aa | ABCC2 | Bombyx mori | [57] | [121] | |||||||||||||||
| Cry1Ab | ABCC2 | Bombyx mori | [58] | [122] | |||||||||||||||
| ABCG1 | Ostrinia furnacalis | [59] | [123] | ||||||||||||||||
| Cry1Ac | ABCC2 | Helicoverpa armigera | [60] | [25] | |||||||||||||||
| ABCC2 | Heliothis virescens | [61] | [124] | ||||||||||||||||
| ABCG1 | Ostrinia furnacalis | [59] | [123] | ||||||||||||||||
| ABCG1 | Plutella xylostella | [62] | [125] | ||||||||||||||||
| ABCC2 | Plutella xylostella | [63] | [126] | ||||||||||||||||
| ABCC2 | Spodoptera exigua | [64] | [127] | ||||||||||||||||
| 8 | 29 | 2 | 1 | 3 | 14 | 3 | 65 | [ | 42 | ] | [54] | ||||||||
| ABCC3 | Spodoptera frugiperda | [65] | [128] | Lygus hesperus | 11 | 6 | 12 | ABCC22 | 1 | 3 | 19 | 11 | 65 | Spodoptera exigua | [64] | [ | [43] | [55] | |
| Cry1Ca | 127 | ] | Lepeophtheirus salmonis | 3 | 4 | 11 | 3 | 1 | 4 | 2 | 5 | 33 | |||||||
| Cry1Fa | ABCC2 | Ostrinia nubilalis | [66] | [129] | [ | 44 | ] | [ | 56 | ] | |||||||||
| Laodelphax striatellux | 2 | 6 | 5 | 2 | 1 | 2 | 14 | 8 | |||||||||||
| Cry2Aa | ABCA2 | Helicoverpa armigera | 40 | [56] | [120] | [ | 45 | ] | [ | 57] | |||||||||
| Bemisia tabaci | 8 | 3 | 6 | 2 | 1 | 3 | 23 | 9 | 55 | ||||||||||
| Cry2Ab | ABCA2 | Helicoverpa armigera | [53] | [68] | [ | 46 | ] | [ | 58 | ] | |||||||||
| Plutella xylostella | 15 | 14 | 21 | 3 | 1 | 3 | 19 | 6 | 82 | [47] | |||||||||
| ABCA2 | Helicoverpa punctigera | [53] | [68] | [ | 48 | ] | [ | 59 | ,60] | ||||||||||
| Manduca sexta | 7 | 9 | 11 | 2 | 1 | 3 | 13 | 3 | 52 | [48] | |||||||||
| ABCA2 | Pectinophora gossypiella | [54] | [118 | [ | 60 | ] | |||||||||||||
| ] | Danaus plexippus | 8 | 16 | 12 | 3 | 1 | 3 | 16 | 3 | 62 | [48] | [60] | |||||||
| Cry3Aa | ABCB1 | Chrysomela tremuela | [67] | [130] | Heliconius melpomene | 10 | 11 | 15 | 2 | 1 | 3 | 17 | 3 | ||||||
| Cry3Bb1 | 62 | [ | 48 | ] | [ | 60 | ] | ||||||||||||
| ABCB1 | Diabrotica virgifera virgifera | [ | 68] | [131] | Helicoverpa armigera | 7 | 11 | 11 | 2 | 1 | 3 | 17 | 2 | 54 | [39][49][50] | [52,61,62] | |||
A Chrysomela tremuela strain survived and reproduced on transgenic poplar trees that expressed a high level of Cry3Aa Bt toxin, and its Cry3Aa resistance was an autosomal recessive trait. Candidate resistance genes were analyzed using midgut transcriptome of larvae, demonstrating that a mutation of the ABCB homolog of P-gp was closely linked to the resistance of Cry3Aa, and named CtABCB1. A 4-bp deletion in CtABCB1 introduced a frame shift with a premature stop codon, resulting in the loss of transporter signature motifs 1 (TpM1) and transmembrane domain 2 (TMD2) [67][130]. A CtABCB1 homologous gene in western corn rootworm (Diabrotica virgifera virgifera) was also shown to be genetically linked to Cry3Bb1 resistance [68][131]. Perhaps the insect ABCB1 transporter is a receptor specific for Cry3 toxin, and structural changes in this transporter may be associated with Cry3 toxin resistance of the insect.
Some ABCC transporters are a functional receptor for more than one Cry1A toxin. The binding of Cry1Ac toxin to ABCC2 on membrane vesicles can lead to membrane perforation. Bretschneider et al. studied the relationship between the ABCC2 of Heliothis virescens and cytotoxicity of three Cry1A toxins (Cry1Aa, Cry1Ab, Cry1Ac) in Spodoptera frugiperda Sf9 cells and found that ABCC2 is the central target of Cry1A toxin action [69][132]. Heterologous expression in D. melanogaster has been used to validate the involvement of ABCC2 in Cry1Ac toxin binding [70][133]. An inactivating mutation, such as mis-splicing of ABCC2 or a mutation causing a single amino acid change, could reduce binding of Cry1Ac toxin, conferring high levels of Bt resistance in the target pest [60][61][71][25,124,134]. In a Cry1Ac-resistant strain of P. xylostella with a 30-bp deletion in exon 20, was expected to cause the removal of the 12th transmembrane (TM) domain and carboxyl terminal of TM12 that is located outside the cell. The mutation may result in the lack of function of a core ATP-binding loop [63][126]. In addition, an ABCC transporter is also the receptor of Cry1Ca and Cry1Fa toxins [64][66][127,129]. A cytotoxicity assay showed that the binding affinity of ABCC transporters to Cry toxin was largely linked to the susceptibility of receptor-expressing cells to Cry toxin and that the extracellular loop (ECL) structures determine the specificity of ABCC to Cry toxins [72][135].
Bt resistance mediated by ABCC transporters involves not only different forms of gene mutations, but also the regulation of ABCC gene expression. The study of Bt resistance in hybrid larvae of B. mori revealed a trans-regulatory mechanism involved in the allele-specific expression of ABCC2 in response to Cry1Ab toxin, which may play an important role in insect Bt resistance [58][122]. The binding of Cry1Ac toxin to ABCC transporters that causes midgut membrane perforation is regulated by a constitutively and transcriptionally activated upstream gene (MAP4K4) in the MAPK signaling pathway [62][125]. In addition, Forkhead box protein A (FOXA) upregulates expression of ABCC2 and ABCC3 genes in Sf9 cells [65][128]. ABCC2 and ABCC3 are important receptors of Cry1Ac toxin, and their expression level has a significant influence on insect Bt resistance; thus, FOXA may be involved in the regulation of insect resistance to Cry1Ac toxin.
ABCG1 (white) gene is one of the most widely studied members of the ABCG protein subfamily. The white protein in P. xylostella is located on the cell membrane, and the expression of Pxwhite gene in the midgut of a Bt-resistant population is significantly lower than in a susceptible population. After RNAi silencing of the midgut Pxwhite gene, the sensitivity of the larvae to Bt-Cry1Ac toxin significantly decreased [73][136]. Genetic linkage analysis confirmed that the decrease was closely linked to the Bt-Cry1Ac resistance of the moth. Downregulated expression of ABCG subfamily genes in Ostrinia furnacalis is also related to its resistance to Cry1Ab and Cry1Ac toxins [59][123].
