You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 3 by Bala S. C. Koritala and Version 2 by Rita Xu.
The circadian clock is a biological clock that regulates processes in cells and whole organs, contributing to dynamic physiology over the 24 hour period.
Intermittent hypoxia (IH) is defined as alternating periods of hypoxia and normoxia. It is associated with multiple respiratory conditions such as chronic obstructive pulmonary disease (COPD) and obstructive sleep apnea (OSA).
- intermittent hypoxia
- circadian rhythms
1. Introduction
Oxygen is an essential element required for the fundamental cellular processes. Insufficient (hypoxia) or excessive (hyperoxia) oxygen alters the functions of molecules, cells, tissues/organs, and can lead to death [1]. Intermittent hypoxia (IH) is a hallmark of multiple respiratory conditions including chronic obstructive pulmonary disease (COPD) and obstructive sleep apnea (OSA) [2][3]. A chronic exposure to IH may be associated with cognitive abnormalities, cardiovascular diseases, as well as metabolic and inflammatory disorders [4]. These comorbidities are also associated with disrupted sleep and misaligned circadian rhythms.
The respiratory patterns of oxygen are controlled by the circadian clock [5]. In turn, oxygen rhythmicity can synchronize the clock through hypoxia-inducible factors (HIFs) [6]. The key transcriptional activators of the hypoxia signaling pathway, HIFs, and the circadian clock components, BMAL1 and CLOCK, belong to the same family of transcription factors [7]. Therefore, HIFs and canonical clock genes can regulate one another and influence circadian rhythms [8]. Despite rodent studies demonstrating that IH impacts gene expression in a time-dependent and tissue-specific manner [9][10], little is known about the impact of IH on the circadian clock. We modeled IH in C57BL6/J mice to represent OSA and better understand the impact of IH on the circadian rhythms of mammalian canonical clock genes. After the exposure to seven days of normoxia or IH, brain (central) and liver (peripheral) tissues were collected and measured the circadian expression of 10 canonical clock genes including Arntl, Clock, Cry1, Cry2, Per1, Per2, Per3, Npas2, Dbp, and Nr1d1.
2. IH Alters Tissue-Specific Circadian Rhythms of Canonical Clock Genes
The central circadian clock in the brain regulates several physiological functions including breathing, hunger, sleep, and body temperature. Compared to other organs, the brain is more sensitive to changes in oxygen levels [11][12]. Therefore, it is important to understand the influence of IH on the clocks in the brain. We measured the differential rhythms of the canonical clock genes between normoxia versus IH (Figure 1A). We observed rhythms in Cry1, Per1, Per2, and Dbp in both conditions. In contrast, the Arntl, Cry2, and Nr1d1 rhythms were altered by the exposure to IH. Arntl and Nr1d1 lost their rhythms, whereas Cry2 rhythms were robust after IH exposure. No detectable rhythms of Clock, Per3, and Npas2 were identified in both conditions. This may be a cause of multiple disparate cell types in the whole brain and/or the sampling resolution in this tissue.


Figure 1. Study design. Six-week-old C57BL/6J male mice were entrained under LD 12:12 h for fourteen days. Animals were then exposed to a seven-day normoxia or intermittent hypoxia (IH) condition. Animals housed under IH received hypoxic events only in the 12 h light phase. After the seventh day of IH exposure, animals from both normoxic and IH conditions were housed under constant exposure to normoxia for two days inircadian rhythms of canonical clock genes. (A) Brain and (B) Liver panels represent the expressions of canonical clock genes over 24 h on the second day of constant darkness. On the second day, central (brain) and peripheral (liver) tissues were collected over 24 h at three-hour intervals. Three animals after exposure to a normoxia (n = 3) were sacirificed at each time point in both conditions. All animals were subjected to ad libitum feeding and stringent temperature control. RNA was purified fromcles; black) and IH (triangles; red). Solid and dashed lines represent rhythmic (p < these0.05) tissues, and the expression of canonical clock genes and non-rhythmic (Arntl,p > Clock, Cry1, Cry2, Per1, Per2, Per3, Npas2, Dbp, Nr1d10.05) was measured using Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR). The data were analyzed through CircaCompare to evaluate differential rhythmicity between conditionsxpression patterns, respectively.
In the liver, there are a higher number of rhythmic genes compared to any other peripheral tissue [13]. The disrupted clock in the liver strongly influences the transcriptional rhythms of other peripheral tissues [14]. Therefore, we measured the circadian rhythms of the canonical clock genes in liver samples (Figure 1B). Eight out of ten canonical genes (Arntl, Cry1, Per1, Per2, Per3, Npas2, Dbp, and Nr1d1) were rhythmic under both conditions. However, Per1 and Nr1d1 were reduced in the amplitude and midline expression in response to IH. Furthermore, the Clock lost its rhythmicity after IH exposure. In addition, the Cry2 rhythm was not detected in both normoxia and IH conditions. This was in line with other large-scale rhythmic expression datasets [15]. Overall, our observations suggested that IH alters the tissue-specific rhythms of canonical clock genes.
3. The Clock in the Liver Is More Stable against IH Compared to the Clock in the Brain
Light–dark cycles and the availability of food are two important zeitgebers (environmental signals) that regulate the mammalian circadian system. The clocks in the brain are directly responsive to light whereas the clocks in the liver are responsive to diet. The functional co-ordination between the brain and the liver plays a significant role in the maintenance of the circadian system for the adaptation and survival of mammals [16]. Therefore, we compared the influence of IH on the clock genes in the liver and the brain using p-value correlation plots (Figure 2). Seven out of ten genes in the brain (Figure 2A) and nine out of ten genes in the liver (Figure 2B) were rhythmic in at least one of the normoxic or IH conditions. Among these rhythmic genes, eight genes in the liver and four genes in the brain did not respond to IH. These results suggested that the circadian rhythms of the clock genes in the liver (Figure 2B) were more stable after exposure to IH compared to brain (Figure 2A).
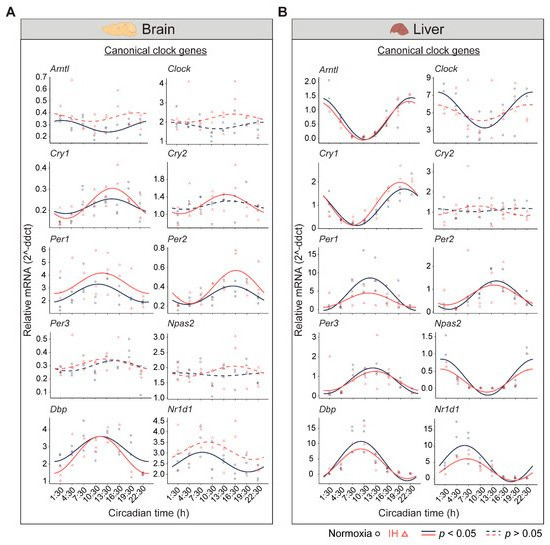
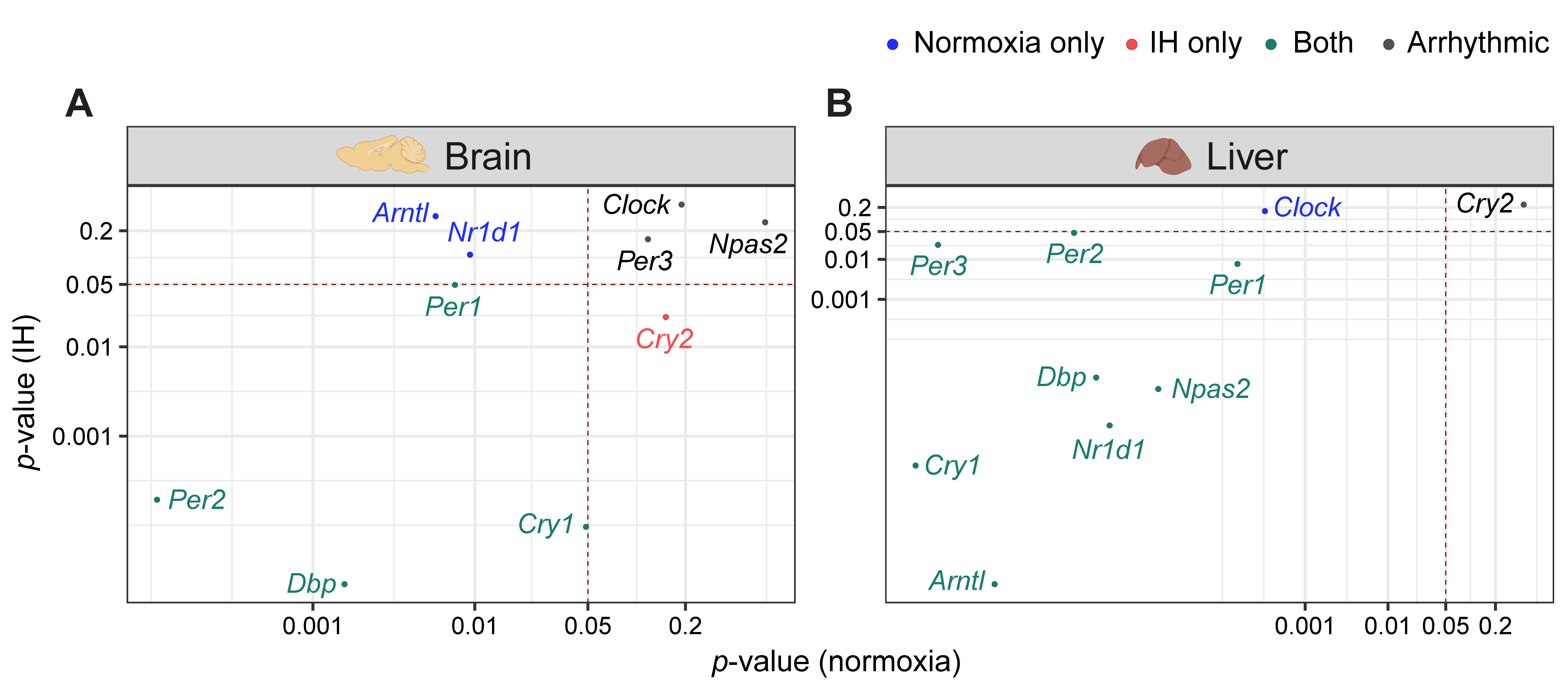


Figure 2. CiIH altercadians rhythms of canonical clic expression of clock genes. (A) BCorrelain and (B) Liver tion panelots represent thrhythmicity p-values expressions ofbetween normoxia vs. IH in brain (A) canod liver (B). Genes rhythmical clock genes over 24 h on the second day of constant darkness after exposure to a normoxia (circles; black) and IH (triangles; red). Solid and dashed lines represent rhythmic (p only in normoxia, only in IH, and in both are represented in blue, red, and cyan, respectively. The number of rhythmic clock genes in both conditions suggests that the circadian clock in the liver is less sensitive to IH compared to the circadian clock <in 0.05) and non-the brain. Arrhythmic (p > 0.05) gexprnession patterns, respectively in both tissues are represented in black.
4. Conclusion
In summary, authors results reinforce the concept that the clock response to IH is tissue-dependent. The organ sensitivity to IH can play a significant role in the alteration of circadian rhythms in the brain. Authors observations also provided insights to potential underlying etiologies of circadian rhythm-associated health conditions.
References
- Wagner, P.D. The biology of oxygen. Eur. Respir. J. 2008, 31, 887–890.
- Tripathi, A.; Melnik, A.V.; Xue, J.; Poulsen, O.; Meehan, M.J.; Humphrey, G.; Jiang, L.; Ackermann, G.; McDonald, D.; Zhou, D.; et al. Intermittent hypoxia and hypercapnia, a hallmark of obstructive sleep apnea, alters the gut microbiome and metabolome. mSystems 2018, 3, e00020-18.
- Zieliński, J. Effects of intermittent hypoxia on pulmonary haemodynamics: Animal models versus studies in humans. Eur. Respir. J. 2005, 25, 173–180.
- Neubauer, J.A. Invited review: Physiological and pathophysiological responses to intermittent hypoxia. J. Appl. Physiol. 2001, 90, 1593–1599.
- Adamovich, Y.; Ladeuix, B.; Sobel, J.; Manella, G.; Neufeld-Cohen, A.; Assadi, M.H.; Golik, M.; Kuperman, Y.; Tarasiuk, A.; Koeners, M.P.; et al. Oxygen and carbon dioxide rhythms are circadian clock controlled and differentially directed by behavioral signals. Cell Metab. 2019, 29, 1092–1103.e3.
- Adamovich, Y.; Ladeuix, B.; Golik, M.; Koeners, M.P.; Asher, G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 2017, 25, 93–101.
- Bersten, D.C.; Sullivan, A.E.; Peet, D.J.; Whitelaw, M.L. bHLH-PAS proteins in cancer. Nat. Rev. Cancer 2013, 13, 827–841.
- O’Connell, E.J.; Martinez, C.-A.; Liang, Y.G.; Cistulli, P.A.; Cook, K.M. Out of breath, out of time: Interactions between HIF and circadian rhythms. Am. J. Physiol. Cell Physiol. 2020, 319, C533–C540.
- Manella, G.; Aviram, R.; Bolshette, N.; Muvkadi, S.; Golik, M.; Smith, D.F.; Asher, G. Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc. Natl. Acad. Sci. USA 2020, 117, 779–786.
- Wu, G.; Lee, Y.Y.; Gulla, E.M.; Potter, A.; Kitzmiller, J.; Ruben, M.D.; Salomonis, N.; Whitsett, J.A.; Francey, L.J.; Hogenesch, J.B.; et al. Short-term exposure to intermittent hypoxia leads to changes in gene expression seen in chronic pulmonary disease. Elife 2021, 10, e63003.
- Casas, A.I.; Geuss, E.; Kleikers, P.W.M.; Mencl, S.; Herrmann, A.M.; Buendia, I.; Egea, J.; Meuth, S.G.; Lopez, M.G.; Kleinschnitz, C.; et al. NOX4-dependent neuronal autotoxicity and BBB breakdown explain the superior sensitivity of the brain to ischemic damage. Proc. Natl. Acad. Sci. USA 2017, 114, 12315–12320.
- Leach, R.M.; Treacher, D.F. Oxygen transport-2. Tissue hypoxia. BMJ 1998, 317, 1370–1373.
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224.
- Manella, G.; Sabath, E.; Aviram, R.; Dandavate, V.; Ezagouri, S.; Golik, M.; Adamovich, Y.; Asher, G. The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nat. Metab. 2021, 3, 829–842.
- Pizarro, A.; Hayer, K.; Lahens, N.F.; Hogenesch, J.B. CircaDB: A da-tabase of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013, 41, D1009–D1013.
- Albrecht, U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron 2012, 74, 246–260.
More
