Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Maheswary Thambirajoo and Version 2 by Dean Liu.
Nanotechnology has become an emerging technology in the medical field and is widely applicable for various clinical applications. The potential use of nanoparticles as antimicrobial agents is greatly explored and taken into consideration as alternative methods to overcome the challenges faced by healthcare workers and patients in preventing infections caused by pathogenic microorganisms.
- nanoparticles
- antimicrobial agents
- antibiotic resistance
1. Introduction
Nanoparticles (NPs) are ultrafine unit particles with one or more dimensions at the nano-size ranging from 1 to 100 nm [1]. There are several factors that influence the physical stability and interactions of NPs with bioactive compounds in vivo. Examples of these factors are the surface morphology, shape, size, and diameter of the particle. The antimicrobial activity of the nanoparticles corresponds to the high surface-area-to-volume ratio. This is because small particles have the largest surface area which increases the interaction with bacteria and improves their antimicrobial activities [2]. In addition, the large surface area to volume also allows the molecules to bind, fuse and integrate the therapeutic agents to the particles [3]. According to Larner and colleagues (2017), NPs often have unexpected visible properties because they are small enough to confine their electrons and produce quantum effects compared to their bulk materials. Thus, NPs confer special optical effects, increase reactivity, and have good stability, but the theory behind these scientific occurrences are still unclear and require further studies for justification [4]. Nanoparticles exhibit unique features and excellent physicochemical characteristics that make them compatible with different biomedical approaches [1][5] including drug delivery systems, radiotherapy, molecular imaging, and treatments for cancer, neurodegenerative disease, lung disease, acquired immune deficiency syndrome (AIDS), and eye disease [6]. Moreover, other industries also highly exploit the use of NPs such as personal care products, electronics devices, buildings, and building materials [7].
NPs are classified based on their constituents and dimensional structure. The constituent is the chemical composition of the material, while the dimensional structure refers to 0D, 1D, 2D and 3D. For example, 0D NPs are holospheres, nanolenses and core–shell quantum dots. 1D NPs consist of nanowires, nanotubes, nanorods and many more. 2D NPs are more to multilayers, films, or plates, while 3D is the crystal form of NPs. The classifications are equally important for researchers as guidelines for safe handling, while the performance of the NPs are based on applications [8]. There is a diverse range of NPs that are currently being studied, namely, carbon-based NPs (carbon nanotubes, fullerene and graphene) [8], metallic-based NPs (silver, gold, copper, iron, arsenic, zinc, nickel, chromium, molybdenum, tantalum, cobalt and antimony) [9][10][11], natural polymeric NPs (chitosan, hyaluronic acid and albumin), synthetic polymeric nanoparticles (poly(glycolic acid), acrylic acid and poly(lactic acid) and dendrimers) [12], lipid-based NPs (solid lipid nanoparticles, nanostructured lipid carriers, lipid–drug conjugates and nanoemulsions) [13] and nanocomposites (metal matrix nanocomposites, polymer matrix nanocomposites and ceramic matrix nanocomposites) [14]. Figure 1 depicts NPs based on classifications and properties. Bottom-up and top-down are the two main techniques applied in the synthesis of NPs, whereby these techniques can be biological, physical, and chemical. The biological techniques use microorganisms such as B. subtilis, sulphate-reducing bacteria S. marcescens, E. coli and B. licheniformis. Both physical and chemical techniques use different substances, such as ammonia, citrate and sodium borohydride, to synthesise NPs [15]. The bottom-up techniques mainly involve a reduction in chemicals, metal vaporisation, electrodes in a chemical solution, light source, high temperature, precipitation, emulsion, sol-gel and many more. The top-down techniques are based on processing a higher scale of nanoparticles to the simplest forms by using a size reduction technique (ball milling), fraction of the metallic atoms on the surface (metal dispersion techniques), and rapid combustion by electrical coil [16].
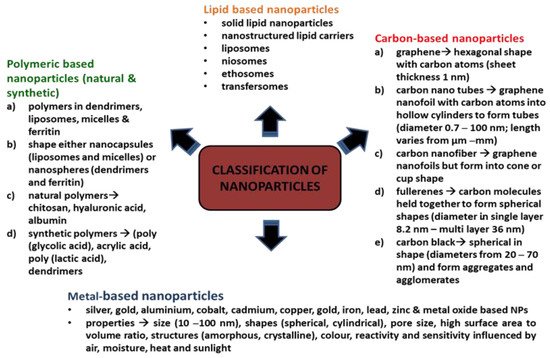
Figure 1. Classifications of nanoparticles.
NPs have gained attention, particularly in antibiotic therapy, as additional substitutes to treat infections caused by various kinds of microorganisms. NPs have emerged as a new promising treatment to combat bacteria or other microorganisms that have developed resistance to antimicrobial drugs, as NPs have excellent bactericidal or microbicidal effects against microbes, such as bacteria, by associating directly with the bacterial cell wall without perforating the cell [17]. In addition, elevated levels of antimicrobial drugs can be initiated and infused at the infected area through NPs as drug delivery vehicles. These NPs have more absorption capacity (i.e., high bioavailability) with longer half-lives and are less likely to cause cell toxicity [11]. Therefore, incorporating NPs with antimicrobial agents or any active moiety of choice can resolve the microbes’ tolerance to drug treatments, maintain optimal antimicrobial concentration, minimise cytotoxicity effects and act as a novel strategy to eradicate microorganisms that cause diseases [11][17].
2. Biofilm Formation and Antibiotic Resistance
It has become a significant challenge for health professionals to treat patients infected by bacteria. This is because prolonged antibiotic or antimicrobial therapies cause bacterial species to become resistant to these treatments since bacteria multiply at a fast rate. The inefficacy of the host immune defence allows for the entry of microbes to colonise and grow [18]. According to Cepas and co-workers (2019), microbes containing adaptable strains or dormant strains, known as “persistent strains”, exhibit antibiotic tolerance and become active once the therapy is withdrawn. These include viruses, bacteria, fungi and parasites. They also added that most of these high-resistance microbe species originated from hospital sites and led to high mortality and morbidity due to the inability of the antibiotic treatments to eliminate these microbes from the infected area. Infections caused by S. aureus have affected people in the US at a cost of 4 billion annually for treatment and management [19]. Emerging infectious diseases (EIDs) are a significant burden on global economics and public health. The incidence of EID events is mostly caused by bacterial species encompassing a large number of drug-resistant microbes [20]. Based on the National Institute of Health, the main causative factor that contributes to pathogenic infections in humans is the formation of biofilm, which is accountable for more than 80% of infections [21]. Commonly, biofilms can be found in chronic wounds, renal infections, cystic fibrosis, severe gum infections, inflammation in the endocardium, inflammation of the meninges, medical-device-associated infections, etc. [21][22].
Other than hospital sites, bacterial biofilm is also present on surfaces in nature, pipeline networks and industrial workplaces in which these biofilms play a dual role, either beneficial or detrimental depending on the conditions of the hosts [23][24]. Biofilms are beneficial as biofertilizers to supply nutrients to crop plants, microbial fuel cells to generate electricity, and to clean up contaminated soil and underground water (bioremediation). Therefore, controlling the formation of biofilms is vital to maintaining biofilms for biotechnological processes and to destroying biofilms for preventing microbes from causing diseases and contaminations [25]. Conventionally, there will be more than one type of microbial species embedded within a biofilm such as in the mouth with 500 different bacterial species in a biofilm [24]. Pathogens, particularly bacteria, form biofilms that function as a barrier to growth, multiply, protect against the host immunity system, resist changes in pH and osmotic concentration or resist drug treatments such as multidrug resistance [26][27]. Biofilm is a clump containing a fraction of bacteria encapsulated with an extracellular polymeric matrix or extracellular polymeric substances (EPS) that attach to the surface area [28]. The EPS is in an aqueous environment containing protein substances and plays a role in maintaining the structural integrity of the biofilm, supplying nutrients, supporting adhesion to surfaces and assisting the intercellular signalling molecules, such as cyclic dimeric guanosine monophosphate (c-di-GMP), that are found in bacterial species within the biofilm. In brief, EPS supports colonisation or re-colonisation of bacteria by adhering to surfaces [29].
Theoretically, there are five stages involved in the formation of a bacterial biofilm [30]. In the first stage, the planktonic bacteria reversibly attach to a suitable surface. In the second stage, the cells begin to connect irreversibly through appendages such as fimbriae, pili, flagella, and lipopolysaccharide (LPS). The third stage involves the cells growing and synthesising EPS containing protein matrix. In the fourth stage, the bacterial cells proliferate and mature to form microcolonies and biofilms. At the final stage, some cells detach or are freed from the biofilm and disperse the contents as planktonic cells to form new biofilms in different locations [30][31]. Figure 2 shows the formation of bacterial biofilms. During biofilm maturation, bacterial cells communicate to other cells within the biofilm or with other microbes through a process known as quorum sensing (QS). This helps the bacteria to keep track of their cell numbers and regulate the expression of quorum-specific genes that facilitate bacterial activity including biofilm formation, virulence, cell mobility, extracting nutrients from other cells and deactivation of the immune system [32]. Ideally, Staphylococcus species is a group of Gram-positive bacteria with a firm biofilm that causes prolonged infections leading to non-wound healing in chronic wounds [33]. S. aureus has the highest resistance followed by S. epidermidis towards antibiotics which are primarily used to prevent hospital-acquired infections caused by these species, specifically with implanted devices. Both species form well-established biofilms, causing more infections compared to other Gram-positive or Gram-negative bacteria. In addition to these bacteria, a few examples of bacterial species that can be found in hospital settings are P. acnes, E. faecalis, S. viridans, E. coli, K. pneumoniae, P. mirabilis and P. aeruginosa [21][34]. In current practice, patients are given more than one type of antibiotic to reduce infections and resolve problems related to drug resistance. The potency of the drugs depends on the patients’ health conditions and requires clinical examinations, antimicrobial sensitivity pattern and aetiological agents to determine the strain causing the infections [20][35].
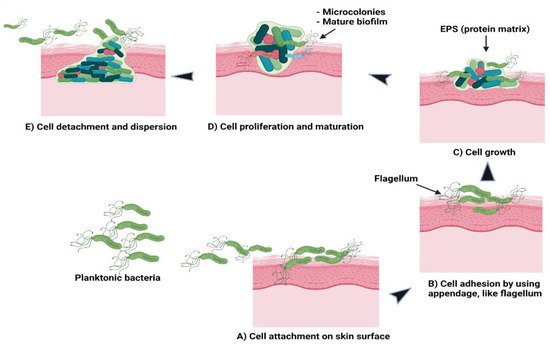
Figure 2. Schematic of (A) bacterial cell attachment to the skin’s surface; (B) irreversible cell attachment through appendages; (C) cells’ growth and EPS synthesis; (D) cells’ proliferation and maturation to form microcolonies and biofilms; (E) cells’ detachment from biofilms to disperse contents as planktonic cells to form new biofilms.
Gradually, after prolonged exposure to antibiotics, microbes are able to mutate their genetic material or modify the antimicrobial agents to escape the effects of these drugs and, at the same time, disable the therapeutic functions of the drugs [36]. The mutation takes place in the genome of the bacteria, particularly the mismatch repair system (MMR) that consists of mutS, mutL, mutH, mutT, mutY, mutM and uvrD, whereby these genes together with DNA gyrase and topoisomerase enzymes can elevate the rate of mutation, inhibiting the antimicrobial agents’ activities [37]. The mutation and modification are accomplished with the help of enzymes synthesised by bacteria based on two mechanisms: (i) alteration of the composition of the bacterial enzymes in such a way as to render the ineffectiveness of the antibiotics while retaining the toxic functions of bacteria and (ii) the antibiotics become weak and dysfunctional after the enzymes change the structural parts of the drugs either by modifying or deactivating them. The modification is achieved by adding several bacterial enzyme groups such as phosphate, adenyl and acetyl. These groups can bind to the target sites of the drugs, altering and developing resistance towards the bacterial activities via inactivation through breaking down of the drugs’ hydrolytic action. Moreover, these enzymes are needed by the bacteria in the production of substances for cellular regulatory processes, cell wall components and nucleic acids. Such examples can be seen in the alteration of bacterial ribosomes by the enzyme of 23S rRNA methyltransferases in Gram-positive bacteria and mutation of DNA gyrase in Gram-negative bacteria. In both mechanisms, macrolides, lincosamides, streptogramins B (for Gram-positive bacteria) and fluoroquinolone antibiotics (for Gram-negative bacteria) are unable to bind to the target site of the bacteria, thus making the drugs ineffective to destroy or kill the bacteria [18][38][39].
NPs utilise a few modes of entry into the biofilm matrix to disrupt the membrane’s cells for cell lysis or cell death. Metals NPs, such as silver, have an active surface area and undergo oxidation to release the Ag+ ions in which these ions fuse to the surface of the bacterial cell wall. The oxidised silver ions that contain more antibacterial properties will penetrate the cell membrane and destroy the cell, which causes the cell to lose its integrity leading to death. [40]. The carbon NPs will elicit electrostatic charges after encountering bacteria and binding to the external part of the cells. The NPs then enter through the damaged cell, destroying DNA replication and draining out the intracellular contents from the cells [41]. Lipid NPs, such as liposomes with positive charges, could damage the biofilm even at low concentrations and retain the capacity of the drugs before assisting the drugs to penetrate the biofilm to kill the cells [40].
Some powerful tools allow the interaction and penetration of NPs into the biofilm such as surface-sensitive techniques, high-resolution microscopies, and synchrotron-based spectroscopies. One example of a surface-sensitive technique is the use of heat produced from gold NPs localised on the surface plasmon resonance. After gold NPs are irradiated, the photon will be absorbed, reflected, or dissipated based on the physical appearance of the NPs. Plasmon is formed after the visible light or infrared radiation is absorbed. Plasmon then changes to hot electrons, which will be balanced with a lattice and release the energy to the neighbouring area. This energy becomes thermal energy and destroys the bacterial cells and biofilms [42]. The second example is the high-resolution microscope. The pH-sensitive polyacrylamide nanosensor (fluorescent nanosensors) that can pierce biofilm developed by P. aeruginosa and S. mutans is able to detect the physiologic pH changes in these biofilms in real-time at the microcolony level. The pH level is elevated while the bacteria start to adapt to acidic conditions until the biofilms are well established. The different gradients of pH can be determined within the individual microcolony, either at the core of the microcolonies or at the edge of the colonies. For example, pH 3.5–4 was observed in the core of the microcolonies and pH 5.5–6 was observed at the edge of the colonies for P. aeruginosa [43]. One of the synchrotron-based spectroscopies was infrared-attenuated total reflectance (IR-ATR) spectroscopy using silver-metal oxide-Teflon-like (CFx) composites to study the antibacterial activity on P. fluorescens biofilm. IR-ATR uses bands to observe biofilm formation or disruption. The bands indicate bacterial adhesion and biofilm expansion. The early stage of the biofilm was observed at the ATR crystal surface followed by EPS and nucleic acids formation. Meanwhile, after modification of the ATR crystal with Ag-CFx, the results demonstrated that in 2 h, the bands related to amide II and EPS were zero, indicating complete elimination of biofilm from the waveguide surface. In contrast to nucleic acids, the bands remained the same throughout the time frame. This could be due to the high antimicrobial activity that is correlated to cell apoptosis and also the reason for the cell membrane destruction of P. fluorescens [44].
Another type of bacterial enzyme which is commonly found in Gram-negative bacte-ria is plasmid-encoded β-Lactamase namely, TEM, SHV, and CTX-M. These enzymes can reduce the effectiveness of drugs such as penicillin, cephalosporins, monobactams and carbapenems by altering the drug binding sites or reduce uptake of drugs due to the changes in the outer membrane of the bacteria. The inactivation of the drugs is based on the hydrolysis process between water molecules from the enzymes and β-lactam rings of the drugs [45]. Figure 3 shows the interactions between antibiotics and bacterial biofilms and how these can lead to antibiotic resistance. Another method the bacteria employ to prevent the entry of antibiotics to the target site is through the efflux pumps, which are the membrane proteins of bacteria. The efflux pumps will export the drugs from the bacterial cell back to the environment, thus increasing their tolerance to antibiotics. P. aeruginosa is an example of bacteria that modulate the transcription regulator of CpxR, which activates efflux pumps and reduces cell membrane permeability to resist drug treatment [46]. Most bacterial species can express efflux pumps either from the same superfamily as a single type of efflux pump or many types from multiple superfamilies [47]. The mechanisms stated above can still operate at the single cell level even after the biofilm formation [48]. The primary functions of antibiotics are to obstruct the essential cellular processes of microbes, specifically, DNA replication, RNA transcription, protein synthesis, impede the synthesis of bacterial cell wall, destruction of cellular membrane, deceleration of cell growth and cell apoptosis [49]. After the formation of biofilm, it is difficult for antimicrobial agents to penetrate deep into the biofilm, and this leads to the drug resistance of bacteria. In addition, bacterial cells that reside deeper in the biofilm tend to multiply slower with lower metabolic activity; hence, they are tolerant to the traditional types of antimicrobial agents [48]. Therefore, coupling NPs with antimicrobial agents could enhance the antimicrobial activity either by piercing into the cell membrane or as drug carriers to directly introduce the NPs to the targeted area for cell disruption [50].
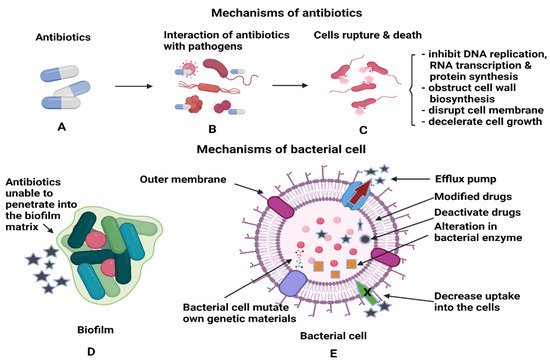
Figure 3. Illustrations of (A,B) the interactions of antibiotics with the pathogens and (C) antibiotics’ activities after penetration into each bacterial cell to kill the cells. (D) Antibiotics inefficiency n killing the bacterial cells, as the cells are protected within the biofilm and (E) the mechanisms utilised by bacterial cells to render the antibiotics’ functions.
References
- Ealias, A.M.; Saravanakumar, M.P. A review on the classification, characterisation, synthesis of nanoparticles and their application. IOP Conf. Ser. Mater. Sci. Eng. 2017, 263, 032019.
- Rudramurthy, G.R.; Swamy, M.K. Potential applications of engineered nanoparticles in medicine and biology: An update. JBIC J. Biol. Inorg. Chem. 2018, 23, 1185–1204.
- Singh, P.; Pandit, S.; Mokkapati, V.; Garg, A.; Ravikumar, V.; Mijakovic, I. Gold Nanoparticles in Diagnostics and Therapeutics for Human Cancer. Int. J. Mol. Sci. 2018, 19, 1979.
- Larner, S.F.; Wang, J.; Goodman, J.; Altman, M.B.O.; Xin, M.; Wang, K. In Vitro Neurotoxicity Resulting from Exposure of Cultured Neural Cells to Several Types of Nanoparticles. J. Cell Death 2017, 10, 1179670717694523.
- Saleh, T.A. Nanomaterials: Classification, properties, and environmental toxicities. Environ. Technol. Innov. 2020, 20, 101067.
- Anderson, S.; Gwenin, V.V.; Gwenin, C.D. Magnetic Functionalized Nanoparticles for Biomedical, Drug Delivery and Imaging Applications. Nanoscale Res. Lett. 2019, 14, 188.
- Mohajerani, A.; Burnett, L.; Smith, J.V.; Kurmus, H.; Milas, J.; Arulrajah, A.; Horpibulsuk, S.; Kadir, A.A. Nanoparticles in Construction Materials and Other Applications, and Implications of Nanoparticle Use. Materials 2019, 12, 3052.
- Prajitha, N.; Athira, S.; Mohanan, P. Bio-interactions and risks of engineered nanoparticles. Environ. Res. 2019, 172, 98–108.
- Amirrah, I.N.; Wee, M.F.M.R.; Tabata, Y.; Idrus, R.B.H.; Nordin, A.; Fauzi, M.B. Antibacterial-Integrated Collagen Wound Dressing for Diabetes-Related Foot Ulcers: An Evidence-Based Review of Clinical Studies. Polymers 2020, 12, 2168.
- Hasan, A.; Morshed, M.; Memic, A.; Hassan, S.; Webster, T.J.; Marei, H. Nanoparticles in tissue engineering: Applications, challenges and prospects. Int. J. Nanomed. 2018, 13, 5637–5655.
- Lakshminarayanan, R.; Ye, E.; Young, D.J.; Li, Z.; Loh, X.J. Recent Advances in the Development of Antimicrobial Nanoparticles for Combating Resistant Pathogens. Adv. Healthc. Mater. 2018, 7, e1701400.
- Palazzolo, S.; Bayda, S.; Hadla, M.; Caligiuri, I.; Corona, G.; Toffoli, G.; Rizzolio, F. The Clinical Translation of Organic Nanomaterials for Cancer Therapy: A Focus on Polymeric Nanoparticles, Micelles, Liposomes and Exosomes. Curr. Med. Chem. 2018, 25, 4224–4268.
- Qi, J.; Zhuang, J.; Lu, Y.; Dong, X.; Zhao, W.; Wu, W. In vivo fate of lipid-based nanoparticles. Drug Discov. Today 2016, 22, 166–172.
- Omanović-Mikličanin, E.; Badnjević, A.; Kazlagić, A.; Hajlovac, M. Nanocomposites: A brief review. Health Technol. 2019, 10, 51–59.
- Rajeshkumar, S.; Veena, P.; Santhiyaa, R.V. Synthesis and Characterization of Selenium Nanoparticles Using Natural Resources and Its Applications. In Exploring the Realms of Nature for Nanosynthesis; Springer: Berlin, Germany, 2018; pp. 63–79.
- Sajid, M.; Płotka-Wasylka, J. Nanoparticles: Synthesis, characteristics, and applications in analytical and other sciences. Microchem. J. 2020, 154, 104623.
- Fernando, S.; Gunasekara, T.; Holton, J. Antimicrobial Nanoparticles: Applications and mechanisms of action. Sri Lankan J. Infect. Dis. 2018, 8, 2.
- Hughes, G.; Webber, M. Novel approaches to the treatment of bacterial biofilm infections. Br. J. Pharmacol. 2017, 174, 2237–2246.
- Cepas, V.; Lopez, V.C.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Marti, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship Between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79.
- Xin, Q.; Shah, H.; Nawaz, A.; Xie, W.; Akram, M.Z.; Batool, A.; Tian, L.; Jan, S.U.; Boddula, R.; Guo, B.; et al. Antibacterial Carbon-Based Nanomaterials. Adv. Mater. 2018, 31, e1804838.
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.-F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067.
- Carbone, A.; Parrino, B.; Cusimano, M.G.; Spanò, V.; Montalbano, A.; Barraja, P.; Schillaci, D.; Cirrincione, G.; Diana, P.; Cascioferro, S. New Thiazole Nortopsentin Analogues Inhibit Bacterial Biofilm Formation. Mar. Drugs 2018, 16, 274.
- Armbruster, C.; Parsek, M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 4317–4319.
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468.
- Berne, C.; Ellison, C.K.; Ducret, A.; Brun, Y.V. Bacterial adhesion at the single-cell level. Nat. Rev. Genet. 2018, 16, 616–627.
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Genet. 2018, 16, 397–409.
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76.
- Sabir, N.; Ikram, A.; Zaman, G.; Satti, L.; Gardezi, A.; Ahmed, A.; Ahmed, P. Bacterial biofilm-based catheter-associated urinary tract infections: Causative pathogens and antibiotic resistance. Am. J. Infect. Control 2017, 45, 1101–1105.
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11.
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11, 928.
- Li, Y.; Li, X.; Hao, Y.; Liu, Y.; Dong, Z.; Li, K. Biological and Physiochemical Methods of Biofilm Adhesion Resistance Control of Medical-Context Surface. Int. J. Biol. Sci. 2021, 17, 1769–1781.
- Azimi, S.; Klementiev, A.D.; Whiteley, M.; Diggle, S.P. Bacterial Quorum Sensing During Infection. Annu. Rev. Microbiol. 2020, 74, 201–219.
- Roy, S.; Santra, S.; DAS, A.; Dixith, S.; Sinha, M.; Ghatak, S.; Ghosh, N.; Banerjee, P.; Khanna, S.; Mathew-Steiner, S.; et al. Staphylococcus aureus Biofilm Infection Compromises Wound Healing by Causing Deficiencies in Granulation Tissue Collagen. Ann. Surg. 2019, 271, 1174–1185.
- De Oliveira, W.F.; Silva, P.; Silva, R.; Silva, G.; Machado, G.; Coelho, L.; Correia, M. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 2018, 98, 111–117.
- Maheswary, T.; Nurul, A.; Fauzi, M. The Insights of Microbes’ Roles in Wound Healing: A Comprehensive Review. Pharmaceutics 2021, 13, 981.
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-Based Nanoparticles as Antimicrobial Agents: An Overview. Nanomaterials 2020, 10, 292.
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39.
- Egorov, A.M.; Ulyashova, M.M.; Rubtsova, M.Y. Bacterial Enzymes and Antibiotic Resistance. Acta Nat. 2018, 10, 33–48.
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month 2020, 66, 100971.
- Lahir, Y.K. Interactions at Interface between Nanomaterial’s and Biofilm: A General Survey. Adv. Clin. Toxicol. 2020, 5.
- Lin, F.; Bao, Y.-W.; Wu, F.-G.; Lin, F.; Bao, Y.-W.; Wu, F.-G. Carbon Dots for Sensing and Killing Microorganisms. C 2019, 5, 33.
- Pihl, M.; Bruzell, E.; Andersson, M. Bacterial biofilm elimination using gold nanorod localised surface plasmon resonance generated heat. Mater. Sci. Eng. C 2017, 80, 54–58.
- Hollmann, B.; Perkins, M.; Chauhan, V.M.; Aylott, J.W.; Hardie, K.R. Fluorescent nanosensors reveal dynamic pH gradients during biofilm formation. NPJ Biofilms Microbiomes 2021, 7, 50.
- Sportelli, M.C.; Tütüncü, E.; Picca, R.A.; Valentini, M.; Kranz, C.; Mizaikoff, B.; Barth, H.; Cioffi, N. Inhibiting P. fluorescens biofilms with fluoropolymer-embedded silver nanoparticles: An in-situ spectroscopic study. Sci. Rep. 2017, 7, 11870.
- González-Bello, C.; Rodríguez, D.; Pernas, M.; Rodríguez, Á.; Colchón-Pierna, E. β-Lactamase Inhibitors to Restore the Efficacy of Antibiotics against Superbugs. J. Med. Chem. 2019, 63, 1859–1881.
- Richardson, L.A. Understanding and overcoming antibiotic resistance. PLoS Biol. 2017, 15, e2003775.
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020.
- Schillaci, D.; Spanò, V.; Parrino, B.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G.; Cascioferro, S. Pharmaceutical Approaches to Target Antibiotic Resistance Mechanisms. J. Med. Chem. 2017, 60, 8268–8297.
- Deliu, I. Antibiotics and antibiotic resistance. Curr Trends Nat. Sci. 2019, 8, 227–232.
- Gupta, A.; Mumtaz, S.; Li, C.-H.; Hussain, I.; Rotello, V.M. Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 2018, 48, 415–427.
More
