Open AccessReviewPeer-Reviewed
Wolf in Sheep’s Clothing: Clostridioides difficile Biofilm as a Reservoir for Recurrent Infections
by Jazmin Meza-Torres 1, Emile Auria 1, Bruno Dupuy1,* and Yannick D. N. Tremblay1,2,*
1 Institut Pasteur, , Université de Paris, UMR-CNRS 2001, Laboratoire Pathogenèse des Bactéries Anaérobies, 25 rue du Docteur Roux, 75724 Paris, France
2 Health Sciences Building, Departement of Biochemistry, Microbiology and Immunology, University of Saskatchewan, 107 Wiggins Rd, Saskatoon, SK S7N 5E5, Canada
*
Auticrobiota ihors to whom correspondence should be addressed.
Academic Editor: Arnaud Bridier
Microorganisms 2021, 9(9), 1922; https://doi.org/10.3390/microorganisms9091922
Received: 10 August 2021 / Revised: 6 September 2021 / Accepted: 7 September 2021 / Published: 10 September 2021
(Thiis article belongs to the Special Issue Microbial Biofilms: Structural Plasticity and Emerging Properties)
Download PDF View PDFBrowse Figurg the ines
Citation Export
Abstriact
The microbiota inhabiting the intestinal tract provide several critical functions to its host. Microorganisms found at the mucosal layer form organized three-dimensional structures which are considered to be biofilms. Their development and functions are influenced by host factors, host-microbe interactions, and microbe-microbe interactions. These structures can dictate the health of their host by strengthening the natural defenses of the gut epithelium or cause disease by exacerbating underlying conditions. Biofilm communities can also block the establishment of pathogens and prevent infectious diseases. Although these biofilms are important for colonization resistance, new data provide evidence that gut biofilms can act as a reservoir for pathogens such as Clostridioides difficile. In this review, we will look at the biofilms of the intestinal tract, their contribution to health and disease, and the factors influencing their formation. We will then focus on the factors contributing to biofilm formation in C. difficile, how these biofilms are formed, and their properties. In the last section, we will look at how the gut microbiota and the gut biofilm influence C. difficile biofilm formation, persistence, and transmission.
- dysbiosis
- mucosal-biofilm
- biofilm
1. Introduction
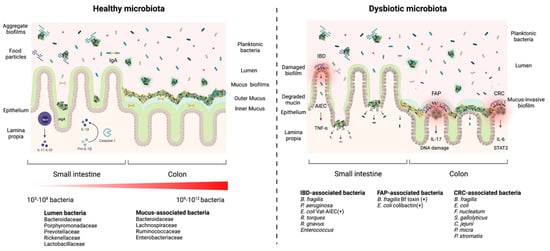
2. Health and Disease: Non-Invasive versus Invasive Gut Microbial Biofilms
3. Diversity of Interactions and Phenotypes in the Gut Biofilm Communities
4. Diverse Gut and Microbiota-Derived Signals Induce Biofilm Formation in Commensal Bacteria and Enteropathogens
The transition from a planktonic state to sessile growth is regulated by multiple steps and regulation cascades, and includes QS-dependent genes, the type IV pili (T4P), and the flagellum [104,105,106][44][45][46]. Biofilm formation is also guided by several environmental signals, which include mechanical signals, nutritional and metabolic cues, inorganic molecules, osmolarity, the presence of antimicrobial molecules, quorum-sensing derived signals, and host-derived signals [107][47]. Bacteria can initiate the transition from a planktonic state to biofilm in vivo to improve their survival against harmful conditions present in the host, to exploit a nutrient rich area that facilitates colonization, or to use the cooperative benefits of multicellular structures [108][48]. Biofilm formation can be controlled by stress response regulators that are activated by different stresses present in the host such as nutrient limitation, iron deprivation, sub-inhibitory concentrations of antibiotics, and osmotic stress [109,110,111][49][50][51]. Specific environmental conditions such as calcium concentration can increase the second messenger c-di-GMP concentrations that could trigger biofilm formation [112][52]. In some cases, biofilm formation is dependent on the nutritional conditions that will trigger metabolic adaptation and thus stimulate biofilm formation [106][46]. In the next section, we will focus on host-derived signals that induce biofilm formation in gut commensal bacteria and enteropathogens.4.1. Host-Derived Factors and Biofilm Formation
4.2. Antibiotics Affecting Biofilm Formation
References
- Eckburg, P.B. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638.
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How Host-Microbial Interactions Shape the Nutrient Environment of the Mammalian Intestine. Annu. Rev. Nutr. 2002, 22, 283–307.
- Hooper, L.V.; Wong, M.H.; Thelin, A.; Hansson, L.; Falk, P.G.; Gordon, J.I. Molecular Analysis of Commensal Host-Microbial Relationships in the Intestine. Science 2001, 291, 881–884.
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 2012, 489, 220–230.
- Motta, J.-P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal Biofilms in Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334.
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of Pathogens and Pathobionts by the Gut Microbiota. Nat. Immunol. 2013, 14, 685–690.
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria Can Protect from Enteropathogenic Infection through Production of Acetate. Nature 2011, 469, 543–547.
- Hammami, R.; Fernandez, B.; Lacroix, C.; Fliss, I. Anti-Infective Properties of Bacteriocins: An Update. Cell. Mol. Life Sci. 2013, 70, 2947–2967.
- McDonald, J.A.K.; Mullish, B.H.; Pechlivanis, A.; Liu, Z.; Brignardello, J.; Kao, D.; Holmes, E.; Li, J.V.; Clarke, T.B.; Thursz, M.R.; et al. Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota. Gastroenterology 2018, 155, 1495–1507.e15.
- Macpherson, A.J.; Geuking, M.B.; McCoy, K.D. Homeland Security: IgA Immunity at the Frontiers of the Body. Trends Immunol. 2012, 33, 160–167.
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The Antibacterial Lectin RegIIIgamma Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science 2011, 334, 255–258.
- Vaishnava, S.; Behrendt, C.L.; Ismail, A.S.; Eckmann, L.; Hooper, L.V. Paneth Cells Directly Sense Gut Commensals and Maintain Homeostasis at the Intestinal Host-Microbial Interface. Proc. Natl. Acad. Sci. USA 2008, 105, 20858–20863.
- Wrzosek, L.; Miquel, S.; Noordine, M.-L.; Bouet, S.; Joncquel Chevalier-Curt, M.; Robert, V.; Philippe, C.; Bridonneau, C.; Cherbuy, C.; Robbe-Masselot, C.; et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii Influence the Production of Mucus Glycans and the Development of Goblet Cells in the Colonic Epithelium of a Gnotobiotic Model Rodent. BMC Biol. 2013, 11, 61.
- Franchi, L.; Kamada, N.; Nakamura, Y.; Burberry, A.; Kuffa, P.; Suzuki, S.; Shaw, M.H.; Kim, Y.-G.; Núñez, G. NLRC4-Driven Production of IL-1β Discriminates between Pathogenic and Commensal Bacteria and Promotes Host Intestinal Defense. Nat. Immunol. 2012, 13, 449–456.
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498.
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e21.
- Collins, J.W.; Keeney, K.M.; Crepin, V.F.; Rathinam, V.A.K.; Fitzgerald, K.A.; Finlay, B.B.; Frankel, G. Citrobacter rodentium: Infection, Inflammation and the Microbiota. Nat. Rev. Microbiol. 2014, 12, 612–623.
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota Organization Is a Distinct Feature of Proximal Colorectal Cancers. Proc. Natl. Acad Sci. USA 2014, 111, 18321–18326.
- Coticchia, J.M.; Sugawa, C.; Tran, V.R.; Gurrola, J.; Kowalski, E.; Carron, M.A. Presence and Density of Helicobacter pylori Biofilms in Human Gastric Mucosa in Patients with Peptic Ulcer Disease. J. Gastrointest. Surg. 2006, 10, 883–889.
- Sigal, M.; Rothenberg, M.E.; Logan, C.Y.; Lee, J.Y.; Honaker, R.W.; Cooper, R.L.; Passarelli, B.; Camorlinga, M.; Bouley, D.M.; Alvarez, G.; et al. Helicobacter pylori Activates and Expands Lgr5(+) Stem Cells Through Direct Colonization of the Gastric Glands. Gastroenterology 2015, 148, 1392–1404.e21.
- Johnson, C.H.; Dejea, C.M.; Edler, D.; Hoang, L.T.; Santidrian, A.F.; Felding, B.H.; Ivanisevic, J.; Cho, K.; Wick, E.C.; Hechenbleikner, E.M.; et al. Metabolism Links Bacterial Biofilms and Colon Carcinogenesis. Cell Metab. 2015, 21, 891–897.
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with Familial Adenomatous polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science 2018, 359, 592–597.
- Chung, L.; Thiele Orberg, E.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis Toxin Coordinates a Pro-Carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e5.
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 2013, 14, 207–215.
- Kumar, R.; Herold, J.L.; Schady, D.; Davis, J.; Kopetz, S.; Martinez-Moczygemba, M.; Murray, B.E.; Han, F.; Li, Y.; Callaway, E.; et al. Streptococcus gallolyticus Subsp. Gallolyticus Promotes Colorectal Tumor Development. PLoS Pathog. 2017, 13, e1006440.
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via Its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206.
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-Resolution Bacterial 16S RRNA Gene Profile Meta-Analysis and Biofilm Status Reveal Common Colorectal Cancer Consortia. NPJ Biofilms Microbiomes 2017, 3, 34.
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-Occurrence of Anaerobic Bacteria in Colorectal Carcinomas. Microbiome 2013, 1, 16.
- Macfarlane, S.; Furrie, E.; Cummings, J.H.; Macfarlane, G.T. Chemotaxonomic Analysis of Bacterial Populations Colonizing the Rectal Mucosa in Patients with Ulcerative Colitis. Clin. Infect. Dis. 2004, 38, 1690–1699.
- Swidsinski, A.; Loening-Baucke, V.; Herber, A. Mucosal Flora in Crohn’s Disease and Ulcerative Colitis—An Overview. J. Physiol. Pharmacol. 2009, 60 (Suppl. S6), 61–71.
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial Organization and Composition of the Mucosal Flora in Patients with Inflammatory Bowel Disease. J. Clin. Microbiol. 2005, 43, 3380–3389.
- Wang, M.; Molin, G.; Ahrné, S.; Adawi, D.; Jeppsson, B. High Proportions of Proinflammatory Bacteria on the Colonic Mucosa in a Young Patient with Ulcerative Colitis as Revealed by Cloning and Sequencing of 16S RRNA Genes. Dig. Dis. Sci. 2007, 52, 620–627.
- Rolhion, N.; Darfeuille-Michaud, A. Adherent-Invasive Escherichia coli in Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2007, 13, 1277–1283.
- Gibold, L.; Garenaux, E.; Dalmasso, G.; Gallucci, C.; Cia, D.; Mottet-Auselo, B.; Faïs, T.; Darfeuille-Michaud, A.; Nguyen, H.T.T.; Barnich, N.; et al. The Vat-AIEC Protease Promotes Crossing of the Intestinal Mucus Layer by Crohn’s Disease-Associated Escherichia coli. Cell Microbiol. 2016, 18, 617–631.
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H.J. Mucolytic Bacteria with Increased Prevalence in IBD Mucosa Augment in vitro Utilization of Mucin by Other Bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428.
- Golińska, E.; Tomusiak, A.; Gosiewski, T.; Więcek, G.; Machul, A.; Mikołajczyk, D.; Bulanda, M.; Heczko, P.B.; Strus, M. Virulence Factors of Enterococcus Strains Isolated from Patients with Inflammatory Bowel Disease. World J. Gastroenterol. 2013, 19, 3562–3572.
- Coyte, K.Z.; Rakoff-Nahoum, S. Understanding Competition and Cooperation within the Mammalian Gut Microbiome. Curr. Biol. 2019, 29, R538–R544.
- Mitri, S.; Richard Foster, K. The Genotypic View of Social Interactions in Microbial Communities. Annu. Rev. Genet. 2013, 47, 247–273.
- Stecher, B.; Robbiani, R.; Walker, A.W.; Westendorf, A.M.; Barthel, M.; Kremer, M.; Chaffron, S.; Macpherson, A.J.; Buer, J.; Parkhill, J.; et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol. 2007, 5, e244.
- Roelofs, K.G.; Coyne, M.J.; Gentyala, R.R.; Chatzidaki-Livanis, M.; Comstock, L.E. Bacteroidales Secreted Antimicrobial Proteins Target Surface Molecules Necessary for Gut Colonization and Mediate Competition In Vivo. mBio 2016, 7, e01055-16.
- Aoki, S.K.; Pamma, R.; Hernday, A.D.; Bickham, J.E.; Braaten, B.A.; Low, D.A. Contact-Dependent Inhibition of Growth in Escherichia coli. Science 2005, 309, 1245–1248.
- Sana, T.G.; Flaugnatti, N.; Lugo, K.A.; Lam, L.H.; Jacobson, A.; Baylot, V.; Durand, E.; Journet, L.; Cascales, E.; Monack, D.M. Salmonella typhimurium Utilizes a T6SS-Mediated Antibacterial Weapon to Establish in the Host Gut. Proc. Natl. Acad. Sci. USA 2016, 113, E5044–E5051.
- Whitney, J.C.; Peterson, S.B.; Kim, J.; Pazos, M.; Verster, A.J.; Radey, M.C.; Kulasekara, H.D.; Ching, M.Q.; Bullen, N.P.; Bryant, D.; et al. A Broadly Distributed Toxin Family Mediates Contact-Dependent Antagonism between Gram-Positive Bacteria. Elife 2017, 6, e26938.
- Barken, K.B.; Pamp, S.J.; Yang, L.; Gjermansen, M.; Bertrand, J.J.; Klausen, M.; Givskov, M.; Whitchurch, C.B.; Engel, J.N.; Tolker-Nielsen, T. Roles of Type IV Pili, Flagellum-Mediated Motility and Extracellular DNA in the Formation of Mature Multicellular Structures in Pseudomonas aeruginosa Biofilms. Environ. Microbiol. 2008, 10, 2331–2343.
- Inhülsen, S.; Aguilar, C.; Schmid, N.; Suppiger, A.; Riedel, K.; Eberl, L. Identification of Functions Linking Quorum Sensing with Biofilm Formation in Burkholderia cenocepacia H111. Microbiologyopen 2012, 1, 225–242.
- Tolker-Nielsen, T. Biofilm Development. Microbiol. Spectr. 2015, 3.
- Karatan, E.; Watnick, P. Signals, Regulatory Networks, and Materials That Build and Break Bacterial Biofilms. Microbiol. Mol. Biol. Rev. 2009, 73, 310–347.
- Jefferson, K.K. What Drives Bacteria to Produce a Biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173.
- Gambino, M.; Cappitelli, F. Mini-Review: Biofilm Responses to Oxidative Stress. Biofouling 2016, 32, 167–178.
- Kang, D.; Kirienko, N.V. Interdependence between Iron Acquisition and Biofilm Formation in Pseudomonas aeruginosa. J. Microbiol. 2018, 56, 449–457.
- Kaplan, J.B. Antibiotic-Induced Biofilm Formation. Int. J. Artif. Organs. 2011, 34, 737–751.
- Chodur, D.M.; Coulter, P.; Isaacs, J.; Pu, M.; Fernandez, N.; Waters, C.M.; Rowe-Magnus, D.A. Environmental Calcium Initiates a Feed-Forward Signaling Circuit That Regulates Biofilm Formation and Rugosity in Vibrio Vulnificus. mBio 2018, 9, e01377-18.
- Hung, D.T.; Zhu, J.; Sturtevant, D.; Mekalanos, J.J. Bile Acids Stimulate Biofilm Formation in Vibrio Cholerae. Mol. Microbiol. 2006, 59, 193–201.
- Koestler, B.J.; Waters, C.M. Intestinal GPS: Bile and Bicarbonate Control Cyclic Di-GMP to Provide Vibrio Cholerae Spatial Cues within the Small Intestine. Gut Microbes 2014, 5, 775–780.
- Köseoğlu, V.K.; Hall, C.P.; Rodríguez-López, E.M.; Agaisse, H. The Autotransporter IcsA Promotes Shigella flexneri Biofilm Formation in the Presence of Bile Salts. Infect. Immun. 2019, 87, e00861-18.
- McKenney, P.T.; Yan, J.; Vaubourgeix, J.; Becattini, S.; Lampen, N.; Motzer, A.; Larson, P.J.; Dannaoui, D.; Fujisawa, S.; Xavier, J.B.; et al. Intestinal Bile Acids Induce a Morphotype Switch in Vancomycin-Resistant Enterococcus That Facilitates Intestinal Colonization. Cell Host Microbe 2019, 25, 695–705.e5.
- Pumbwe, L.; Skilbeck, C.A.; Nakano, V.; Avila-Campos, M.J.; Piazza, R.M.F.; Wexler, H.M. Bile Salts Enhance Bacterial Co-Aggregation, Bacterial-Intestinal Epithelial Cell Adhesion, Biofilm Formation and Antimicrobial Resistance of Bacteroides Fragilis. Microb. Pathog. 2007, 43, 78–87.
- Kelly, S.M.; Lanigan, N.; O’Neill, I.J.; Bottacini, F.; Lugli, G.A.; Viappiani, A.; Turroni, F.; Ventura, M.; van Sinderen, D. Bifidobacterial Biofilm Formation Is a Multifactorial Adaptive Phenomenon in Response to Bile Exposure. Sci. Rep. 2020, 10, 11598.
- López, M.; Blasco, L.; Gato, E.; Perez, A.; Fernández-Garcia, L.; Martínez-Martinez, L.; Fernández-Cuenca, F.; Rodríguez-Baño, J.; Pascual, A.; Bou, G.; et al. Response to Bile Salts in Clinical Strains of Acinetobacter baumannii Lacking the AdeABC Efflux Pump: Virulence Associated with Quorum Sensing. Front. Cell Infect. Microbiol. 2017, 7, 143.
- Wang, X.; Wang, Y.; Ling, N.; Shen, Y.; Zhang, D.; Liu, D.; Ou, D.; Wu, Q.; Ye, Y. Roles of TolC on Tolerance to Bile Salts and Biofilm Formation in Cronobacter malonaticus. J. Dairy Sci. 2021, 104, 9521–9531.
- Ambalam, P.; Kondepudi, K.K.; Nilsson, I.; Wadström, T.; Ljungh, A. Bile Enhances Cell Surface Hydrophobicity and Biofilm Formation of Bifidobacteria. Appl. Biochem. Biotechnol. 2014, 172, 1970–1981.
- Bechon, N.; Mihajlovic, J.; Lopes, A.-A.; Vendrell-Fernández, S.; Deschamps, J.; Briandet, R.; Sismeiro, O.; Martin-Verstraete, I.; Dupuy, B.; Ghigo, J.-M. Bacteroides thetaiotaomicron Uses a Widespread Extracellular DNase to Promote Bile-Dependent Biofilm Formation. bioRxiv 2021.
- Bollinger, R.R.; Everett, M.L.; Palestrant, D.; Love, S.D.; Lin, S.S.; Parker, W. Human Secretory Immunoglobulin A May Contribute to Biofilm Formation in the Gut. Immunology 2003, 109, 580–587.
- Everett, M.L.; Palestrant, D.; Miller, S.E.; Bollinger, R.R.; Parker, W. Immune Exclusion and Immune Inclusion: A New Model of Host-Bacterial Interactions in the Gut. Clin. Appl. Immunol. Rev. 2004, 4, 321–332.
- Popowska, M.; Krawczyk-Balska, A.; Ostrowski, R.; Desvaux, M. InlL from Listeria monocytogenes Is Involved in Biofilm Formation and Adhesion to Mucin. Front. Microbiol. 2017, 8, 660.
- Tu, Q.V.; McGuckin, M.A.; Mendz, G.L. Campylobacter jejuni Response to Human Mucin MUC2: Modulation of Colonization and Pathogenicity Determinants. J. Med. Microbiol. 2008, 57, 795–802.
- Dwivedi, R.; Nothaft, H.; Garber, J.; Xin Kin, L.; Stahl, M.; Flint, A.; van Vliet, A.H.M.; Stintzi, A.; Szymanski, C.M. L-Fucose Influences Chemotaxis and Biofilm Formation in Campylobacter jejuni. Mol. Microbiol. 2016, 101, 575–589.
- Motta, J.-P.; Flannigan, K.L.; Agbor, T.A.; Beatty, J.K.; Blackler, R.W.; Workentine, M.L.; Da Silva, G.J.; Wang, R.; Buret, A.G.; Wallace, J.L. Hydrogen Sulfide Protects from Colitis and Restores Intestinal Microbiota Biofilm and Mucus Production. Inflamm. Bowel. Dis. 2015, 21, 1006–1017.
- Feraco, D.; Blaha, M.; Khan, S.; Green, J.M.; Plotkin, B.J. Host Environmental Signals and Effects on Biofilm Formation. Microb. Pathog. 2016, 99, 253–263.
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria-Host Communication: The Language of Hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956.
- Plotkin, B.J.; Wu, Z.; Ward, K.; Nadella, S.; Green, J.M.; Rumnani, B. Effect of Human Insulin on the Formation of Catheter-Associated E. coli biofilms. OJU 2014, 4, 49–56.
- Klosowska, K.; Plotkin, B.J. Human Insulin Modulation of Escherichia coli Adherence and Chemotaxis. Am. J. Infect. Dis. 2006, 2, 197–200.
- Scardaci, R.; Varese, F.; Manfredi, M.; Marengo, E.; Mazzoli, R.; Pessione, E. Enterococcus faecium NCIMB10415 Responds to Norepinephrine by Altering Protein Profiles and Phenotypic Characters. J. Proteom. 2021, 231, 104003.
- Cambronel, M.; Nilly, F.; Mesguida, O.; Boukerb, A.M.; Racine, P.-J.; Baccouri, O.; Borrel, V.; Martel, J.; Fécamp, F.; Knowlton, R.; et al. Influence of Catecholamines (Epinephrine/Norepinephrine) on Biofilm Formation and Adhesion in Pathogenic and Probiotic Strains of Enterococcus faecalis. Front. Microbiol. 2020, 11, 1501.
- Hiller, C.C.; Lucca, V.; Carvalho, D.; Borsoi, A.; Borges, K.A.; Furian, T.Q.; do Nascimento, V.P. Influence of Catecholamines on Biofilm Formation by Salmonella enteritidis. Microb. Pathog. 2019, 130, 54–58.
- Maestre, J.R.; Aguilar, L.; Mateo, M.; Giménez, M.-J.; Méndez, M.-L.; Alou, L.; Granizo, J.-J.; Prieto, J. In vitro Interference of Tigecycline at Subinhibitory Concentrations on Biofilm Development by Enterococcus faecalis. J. Antimicrob. Chemother. 2012, 67, 1155–1158.
- Ozma, M.A.; Khodadadi, E.; Rezaee, M.A.; Kamounah, F.S.; Asgharzadeh, M.; Ganbarov, K.; Aghazadeh, M.; Yousefi, M.; Pirzadeh, T.; Kafil, H.S. Induction of Proteome Changes Involved in Biofilm Formation of Enterococcus faecalis in Response to Gentamicin. Microb. Pathog. 2021, 157, 105003.
- Yu, W.; Hallinen, K.M.; Wood, K.B. Interplay between Antibiotic Efficacy and Drug-Induced Lysis Underlies Enhanced Biofilm Formation at Subinhibitory Drug Concentrations. Antimicrob. Agents Chemother. 2018, 62, e01603-17.
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside Antibiotics Induce Bacterial Biofilm Formation. Nature 2005, 436, 1171–1175.
- Sun, F.; Yuan, Q.; Wang, Y.; Cheng, L.; Li, X.; Feng, W.; Xia, P. Sub-Minimum Inhibitory Concentration Ceftazidime Inhibits Escherichia coli Biofilm Formation by Influencing the Levels of the IbpA Gene and Extracellular Indole. J. Chemother. 2020, 32, 7–14.
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020.
- Schembri, M.A.; Kjaergaard, K.; Klemm, P. Global Gene Expression in Escherichia coli Biofilms. Mol. Microbiol. 2003, 48, 253–267.
- Xavier, K.B.; Bassler, B.L. Regulation of Uptake and Processing of the Quorum-Sensing Autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005, 187, 238–248.
- Bay, D.C.; Stremick, C.A.; Slipski, C.J.; Turner, R.J. Secondary Multidrug Efflux Pump Mutants Alter Escherichia coli Biofilm Growth in the Presence of Cationic Antimicrobial Compounds. Res. Microbiol. 2017, 168, 208–221.
- Matsumura, K.; Furukawa, S.; Ogihara, H.; Morinaga, Y. Roles of Multidrug Efflux Pumps on the Biofilm Formation of Escherichia coli K-12. Biocontrol. Sci. 2011, 16, 69–72.
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.V.; Webber, M.A. Loss of or Inhibition of All Multidrug Resistance Efflux Pumps of Salmonella enterica Serovar Typhimurium Results in Impaired Ability to Form a Biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417.
- Teteneva, N.A.; Mart’yanov, S.V.; Esteban-López, M.; Kahnt, J.; Glatter, T.; Netrusov, A.I.; Plakunov, V.K.; Sourjik, V. Multiple Drug-Induced Stress Responses Inhibit Formation of Escherichia coli Biofilms. Appl. Environ. Microbiol. 2020, 86, e01113-20.
