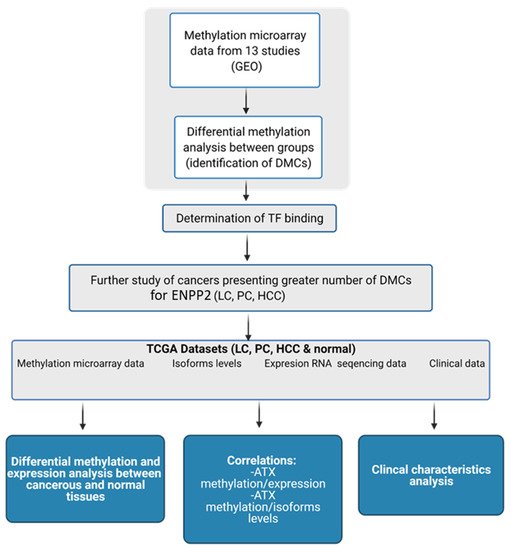
Autotaxin (ATX) encoded by Ectonucleotide Pyrophosphatase/Phosphodiesterase 2 (ENPP2) is a key enzyme in Lysophosphatidic Acid (LPA) synthesis implicated in cancer. Although its aberrant expression has been reported, ENPP2 methylation profiles in health and malignancy are not described. We examined in silico the methylation of ENPP2 analyzing publicly available methylome datasets, to identify Differentially Methylated CpGs (DMCs) which were then correlated with expression at gene and isoform levels. Significance indication was set to be FDR corrected p-value < 0.05. Healthy tissues presented methylation in all gene body CGs and lower levels in Promoter Associated (PA) regions, whereas in the majority of the tumors examined (HCC, melanoma, CRC, LC and PC) the methylation pattern was reversed. DMCs identified in the promoter were located in sites recognized by multiple transcription factors, suggesting involvement in gene expression. Alterations in methylation were correlated to an aggressive phenotype in cancer cell lines. In prostate and lung adenocarcinomas, increased methylation of PA CGs was correlated to decreased ENPP2 mRNA expression and to poor prognosis parameters. Collectively, our results corroborate that methylation is an active level of ATX expression regulation in cancer. Our study provides an extended description of the methylation status of ENPP2 in health and cancer and points out specific DMCs of value as prognostic biomarkers.
- autotaxin
- ENPP2
- methylation
- cancer
- expression
- regulation
1. Introduction
2. Current Insights
References
- Stefan, C.; Jansen, S.; Bollen, M. NPP-type ectophosphodiesterases: Unity in diversity. Trends Biochem. Sci. 2005, 30, 542–550.
- Tanaka, M.; Okudaira, S.; Kishi, Y.; Ohkawa, R.; Iseki, S.; Ota, M.; Noji, S.; Yatomi, Y.; Aoki, J.; Arai, H. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J. Biol. Chem. 2006, 281, 25822–25830.
- Ninou, I.; Magkrioti, C.; Aidinis, V. Autotaxin in Pathophysiology and Pulmonary Fibrosis. Front. Med. 2018, 5, 180.
- Magkrioti, C.; Galaris, A.; Kanellopoulou, P.; Stylianaki, E.A.; Kaffe, E.; Aidinis, V. Autotaxin and chronic inflammatory diseases. J. Autoimmun. 2019, 104, 102327.
- Peyruchaud, O.; Saier, L.; Leblanc, R. Autotaxin Implication in Cancer Metastasis and Autoimunne Disorders: Functional Implication of Binding Autotaxin to the Cell Surface. Cancers 2019, 12, 105.
- Benesch, M.G.; Tang, X.; Maeda, T.; Ohhata, A.; Zhao, Y.Y.; Kok, B.P.; Dewald, J.; Hitt, M.; Curtis, J.M.; McMullen, T.P.; et al. Inhibition of autotaxin delays breast tumor growth and lung metastasis in mice. FASEB J. 2014, 28, 2655–2666.
- Memet, I.; Tsalkidou, E.; Tsaroucha, A.K.; Lambropoulou, M.; Chatzaki, E.; Trypsianis, G.; Schizas, D.; Pitiakoudis, M.; Simopoulos, C. Autotaxin Expression in Hepatocellular Carcinoma. J. Investig. Surg. 2018, 31, 359–365.
- Yang, S.Y.; Lee, J.; Park, C.G.; Kim, S.; Hong, S.; Chung, H.C.; Min, S.K.; Han, J.W.; Lee, H.W.; Lee, H.Y. Expression of autotaxin (NPP-2) is closely linked to invasiveness of breast cancer cells. Clin. Exp. Metastasis 2002, 19, 603–608.
- Oikonomou, N.; Mouratis, M.A.; Tzouvelekis, A.; Kaffe, E.; Valavanis, C.; Vilaras, G.; Karameris, A.; Prestwich, G.D.; Bouros, D.; Aidinis, V. Pulmonary autotaxin expression contributes to the pathogenesis of pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2012, 47, 566–574.
- Lee, H.Y.; Murata, J.; Clair, T.; Polymeropoulos, M.H.; Torres, R.; Manrow, R.E.; Liotta, L.A.; Stracke, M.L. Cloning, chromosomal localization, and tissue expression of autotaxin from human teratocarcinoma cells. Biochem. Biophys. Res. Commun. 1996, 218, 714–719.
- Brisbin, A.G.; Asmann, Y.W.; Song, H.; Tsai, Y.Y.; Aakre, J.A.; Yang, P.; Jenkins, R.B.; Pharoah, P.; Schumacher, F.; Conti, D.V.; et al. Meta-analysis of 8q24 for seven cancers reveals a locus between NOV and ENPP2 associated with cancer development. BMC Med. Genet. 2011, 12, 156.
- Giganti, A.; Rodriguez, M.; Fould, B.; Moulharat, N.; Cogé, F.; Chomarat, P.; Galizzi, J.-P.; Valet, P.; Saulnier-Blache, J.-S.; Boutin, J.A.; et al. Murine and Human Autotaxin α, β, and γ Isoforms: Gene organization, tissue distribution, and biochemical characterization. J. Biol. Chem. 2008, 283, 7776–7789.
- Parris, T.Z.; Kovács, A.; Hajizadeh, S.; Nemes, S.; Semaan, M.; Levin, M.; Karlsson, P.; Helou, K. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogenesis 2014, 3, e95.
- Razin, A.; Cedar, H. DNA methylation and gene expression. Microbiol. Rev. 1991, 55, 451–458.
- Nikolaidis, C.; Nena, E.; Panagopoulou, M.; Balgkouranidou, I.; Karaglani, M.; Chatzaki, E.; Agorastos, T.; Constantinidis, T.C. PAX1 methylation as an auxiliary biomarker for cervical cancer screening: A meta-analysis. Cancer Epidemiol. 2015, 39, 682–686.
- Panagopoulou, M.; Esteller, M.; Chatzaki, E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers 2021, 13, 728.
- Panagopoulou, M.; Karaglani, M.; Manolopoulos, V.G.; Iliopoulos, I.; Tsamardinos, I.; Chatzaki, E. Deciphering the Methylation Landscape in Breast Cancer: Diagnostic and Prognostic Biosignatures through Automated Machine Learning. Cancers 2021, 13, 1677.
- Panagopoulou, M.; Cheretaki, A.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Amarantidis, K.; Xenidis, N.; Kakolyris, S.; Baritaki, S.; Chatzaki, E. Methylation Status of Corticotropin-Releasing Factor (CRF) Receptor Genes in Colorectal Cancer. J. Clin. Med. 2021, 10, 2680.
- Panagopoulou, M.; Karaglani, M.; Balgkouranidou, I.; Biziota, E.; Koukaki, T.; Karamitrousis, E.; Nena, E.; Tsamardinos, I.; Kolios, G.; Lianidou, E.; et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2019, 38, 3387–3401.
- Panagopoulou, M.; Lambropoulou, M.; Balgkouranidou, I.; Nena, E.; Karaglani, M.; Nicolaidou, C.; Asimaki, A.; Konstantinidis, T.; Constantinidis, T.C.; Kolios, G.; et al. Gene promoter methylation and protein expression of BRMS1 in uterine cervix in relation to high-risk human papilloma virus infection and cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317697557.
- Udomsinprasert, W.; Kitkumthorn, N.; Mutirangura, A.; Chongsrisawat, V.; Poovorawan, Y.; Honsawek, S. Association between Promoter Hypomethylation and Overexpression of Autotaxin with Outcome Parameters in Biliary Atresia. PLoS ONE 2017, 12, e0169306.
- Liu, X.; Peng, Y.; Wang, J. Integrative analysis of DNA methylation and gene expression profiles identified potential breast cancer-specific diagnostic markers. Biosci. Rep. 2020, 40.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Perrakis, A.; Moolenaar, W.H. Autotaxin: Structure-function and signaling. J. Lipid Res. 2014, 55, 1010–1018.
- Fuss, B.; Baba, H.; Phan, T.; Tuohy, V.K.; Macklin, W.B. Phosphodiesterase I, a novel adhesion molecule and/or cytokine involved in oligodendrocyte function. J. Neurosci. 1997, 17, 9095–9103.
- Kanda, H.; Newton, R.; Klein, R.; Morita, Y.; Gunn, M.D.; Rosen, S.D. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nat. Immunol. 2008, 9, 415–423.
- Nema, R.; Shrivastava, A.; Kumar, A. Prognostic role of lipid phosphate phosphatases in non-smoker, lung adenocarcinoma patients. Comput. Biol. Med. 2021, 129, 104141.
- Wang, S.C.; Liao, L.M.; Ansar, M.; Lin, S.Y.; Hsu, W.W.; Su, C.M.; Chung, Y.M.; Liu, C.C.; Hung, C.S.; Lin, R.K. Automatic Detection of the Circulating Cell-Free Methylated DNA Pattern of GCM2, ITPRIPL1 and CCDC181 for Detection of Early Breast Cancer and Surgical Treatment Response. Cancers 2021, 13, 1375.
- Nouh, M.A.; Wu, X.X.; Okazoe, H.; Tsunemori, H.; Haba, R.; Abou-Zeid, A.M.; Saleem, M.D.; Inui, M.; Sugimoto, M.; Aoki, J.; et al. Expression of autotaxin and acylglycerol kinase in prostate cancer: Association with cancer development and progression. Cancer Sci. 2009, 100, 1631–1638.
- Magkrioti, C.; Oikonomou, N.; Kaffe, E.; Mouratis, M.A.; Xylourgidis, N.; Barbayianni, I.; Megadoukas, P.; Harokopos, V.; Valavanis, C.; Chun, J.; et al. The Autotaxin-Lysophosphatidic Acid Axis Promotes Lung Carcinogenesis. Cancer Res. 2018, 78, 3634–3644.
- Wu, J.M.; Xu, Y.; Skill, N.J.; Sheng, H.; Zhao, Z.; Yu, M.; Saxena, R.; Maluccio, M.A. Autotaxin expression and its connection with the TNF-alpha-NF-kappaB axis in human hepatocellular carcinoma. Mol. Cancer 2010, 9, 71.
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973.
- Chen, G.; Gharib, T.G.; Huang, C.C.; Taylor, J.M.; Misek, D.E.; Kardia, S.L.; Giordano, T.J.; Iannettoni, M.D.; Orringer, M.B.; Hanash, S.M.; et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell. Proteom. MCP 2002, 1, 304–313.
- Lemée, J.M.; Clavreul, A.; Aubry, M.; Com, E.; de Tayrac, M.; Mosser, J.; Menei, P. Integration of transcriptome and proteome profiles in glioblastoma: Looking for the missing link. BMC Mol. Biol. 2018, 19, 13.
- Arcos-Montoya, D.; Wegman-Ostrosky, T.; Mejía-Pérez, S.; De la Fuente-Granada, M.; Camacho-Arroyo, I.; García-Carrancá, A.; Velasco-Velázquez, M.A.; Manjarrez-Marmolejo, J.; González-Arenas, A. Progesterone Receptor Together with PKCα Expression as Prognostic Factors for Astrocytomas Malignancy. OncoTargets Ther. 2021, 14, 3757–3768.
- Tate, P.H.; Bird, A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993, 3, 226–231.
- Farina, A.R.; Cappabianca, L.; Ruggeri, P.; Di Ianni, N.; Ragone, M.; Merolle, S.; Sano, K.; Stracke, M.L.; Horowitz, J.M.; Gulino, A.; et al. Constitutive autotaxin transcription by Nmyc-amplified and non-amplified neuroblastoma cells is regulated by a novel AP-1 and SP-mediated mechanism and abrogated by curcumin. FEBS Lett. 2012, 586, 3681–3691.
- Zhang, X.; Li, M.; Yin, N.; Zhang, J. The Expression Regulation and Biological Function of Autotaxin. Cells 2021, 10, 939.
- Héberlé, É.; Bardet, A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019, 63, 727–741.
- Chen, M.; O’Connor, K.L. Integrin alpha6beta4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene 2005, 24, 5125–5130.
- Braeuer, R.R.; Zigler, M.; Kamiya, T.; Dobroff, A.S.; Huang, L.; Choi, W.; McConkey, D.J.; Shoshan, E.; Mobley, A.K.; Song, R.; et al. Galectin-3 contributes to melanoma growth and metastasis via regulation of NFAT1 and autotaxin. Cancer Res. 2012, 72, 5757–5766.
- Sioletic, S.; Czaplinski, J.; Hu, L.; Fletcher, J.A.; Fletcher, C.D.; Wagner, A.J.; Loda, M.; Demetri, G.D.; Sicinska, E.T.; Snyder, E.L. c-Jun promotes cell migration and drives expression of the motility factor ENPP2 in soft tissue sarcomas. J. Pathol. 2014, 234, 190–202.
- Benesch, M.G.; Tang, X.; Venkatraman, G.; Bekele, R.T.; Brindley, D.N. Recent advances in targeting the autotaxin-lysophosphatidate-lipid phosphate phosphatase axis in vivo. J. Biomed. Res. 2016, 30, 272–284.
- Barbayianni, E.; Kaffe, E.; Aidinis, V.; Kokotos, G. Autotaxin, a secreted lysophospholipase D, as a promising therapeutic target in chronic inflammation and cancer. Prog. Lipid Res. 2015, 58, 76–96.
