You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Dean Liu and Version 1 by Ian Macreadie.
The high transmissibility of Delta and Lambda variants has raised the need to determine the reason for the infectivity of these variants. Since the Delta variant first emerged in India in June 2020, the global incidence of the Delta variant has made it a VOC. The Delta variant is 60% more infectious than the original Wuhan SARS-CoV-2 strain. Similarly, Lambda, a VOI, was first reported in Peru in December, 2020 and currently, its diverse mutations and increased frequency in over 29 countries has raised more attention.
- SARS-CoV2
- Delta variant
- Lambda variant
1. Introduction
Compared to SARS-CoV and MERS-CoV, the SARS-CoV-2 genome has 82% sequence identity overall, and over 90% sequence identity for structural proteins and essential enzymes [9][1]. According to the NCBI annotation, (NC_045512), SARS-CoV-2 has 14 functional open reading frames (ORFs), which encode up to 31 proteins, shown schematically in Figure 1 [3,10][2][3]. The first ORFs, ORF1a and ORF1b, are the longest ORFs and occupy more than two thirds of the whole genome [3,11][2][4]. Polyproteins 1a and 1b are encoded by ORFs 1a and 1b. These two polyproteins are cleaved with the aid of proteases such as Mpro to produce 16 non-structural proteins (NSPs) [9,12,13][1][5][6]. Mpro, the main protease of the coronaviruses, is responsible for viral replication and transcription and is described in detail in previous studies [12,13][5][6]. The NSPs include regulators of the viral lifecycle, which play a crucial role in the replication and pathogenicity of the virus [9,14][1][7]. The mechanism of viral invasion and protein functions was reviewed previously [15,16,17][8][9][10]. Downstream in the RNA are 11 accessory proteins encoded by ORFs, whose order is ORF3a, ORF3b, ORF3c, ORF3d, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c and ORF10, respectively [18][11]. They interact with four structural proteins, but their roles remain unknown [19][12]. The four structural proteins are spike surface glycoprotein (S), small envelope protein (E), membrane or matrix protein (M), and nucleocapsid protein (N) [20][13]. Among the structural proteins, S protein plays a vital role in the pathogenicity and transmission of the virus (Figure 1).
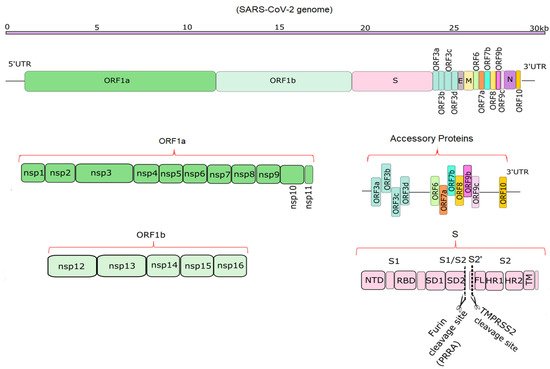
Figure 1. Schematic diagrams of the SARS-CoV-2 genome and its translation products. The top part shows the ~29.9 kb genome [1][14]. The second portion shows the overall position of structural, non-structural and accessory proteins in 5′ to 3′ order [3,21][2][15]. The bottom half depicts the non-structural proteins encoded by ORF1a (NSP1-11) and ORF1b (NSP12-16), 11 accessory proteins encoded by ORF3-ORF10, and the structure of S, which has S1 and S2 domains [21][15]. Furin and TMPRSS2 cleavage sites (PRRA and S2′ sites) are depicted in the S protein [22][16].
2. Why Delta and Lamba Variants Are More Pathogenic
Apart from the toxicity of the S protein, the S1 domain is highly mutable and may not be a suitable candidate for drug targeting [29][17]. In contrast, cell-mediated immune responses to the S protein are mainly elicited to the S2 domain [42][18]. This also contains two heptad repeat regions (HR1 and HR2), which provide the S2 domain as a target of NAbs and vaccine candidate [43][19]. Mutations in spike protein may lead to increased affinity between the RBD and the ACE2 [30,44][20][21]. Among the 17 mutations of the Delta spike protein, the L452R and T478K mutations are notable because amino acid substitutions at positions 452 and 478 have increased affinity of the spike protein for ACE2 receptors. Thus, they increase the capability of the virus to evade a host’s immune response [43][19]. There is a specific mutation, P681R, found within the furin cleavage site of the Delta variant [45][22]. An initial study in Delta demonstrates the increased processing of spike protein, compared with the original SARS-CoV-2 [46][23]. In this regard, higher fusion activity and syncytium formation has been reported in the Delta variant, compared with previous lineages [44][21]. Similarly, there are 19 mutations in the Lambda variant, including a unique deletion of 7 amino acids at positions 246–253, plus the substitution D253N located at the NTD of the S protein (Figure 2), the antigenic supersite, raising the prospect of more transmissibility and more infection in the Lambda variant [8][24]. However, T76I, F490S and L452Q substitutions in Lambda also occur in the RBD of spike protein [47][25]. As seen in Figure 2, the most significant changes within the RBD for the two variants are L452R and T478K in Delta, and T76I, F490S and L452Q in Lambda. Generally, mutations that occur in the RBD and the N-terminal domain allow the virus to evade the host immune response. Consequently, the Delta and Lambda variants appears to have decreased sensitivity to antibodies [6,48][26][27] elicited by the current vaccine spike protein. The molecular structure, important amino acid changes and protein–protein binding of spike with ACE2 for Delta and Lambda variants are represented in Figure 2.
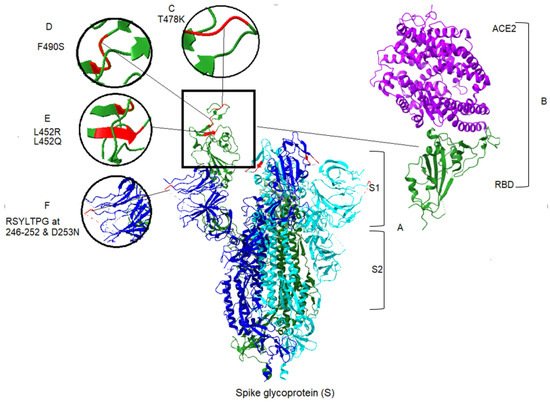
Figure 2. Spike protein and associated mutations in S1 domain for Delta and Lambda variants of SARS-CoV-2, and ACE2 binding to the S protein. (A) The crystal structure of S protein domains (S1 and S2) in the active conformation of the RBD of spike protein (Chain A). The PDB format was obtained from the RCSB protein data bank (PDB ID: 7DX5) [49][28]. Homology modeling was performed, using UCSF ChimeraX version 1.2.5 (2021-05-24). ACE2 was removed from the structure in the central panel [50][29]. (B) Right panel, the partial conformation of the structure of RBD of the spike protein (Chain F), shown in a green color, in complex with ACE2 (Chain B) in purple color, retrieved from the RCSB protein data bank (PDB ID: 6vw1) to demonstrate the RBD binding to hACE2 [51][30]. Structural modeling and point mutations were carried out, using UCSF ChimeraX. Other chains and molecules were removed. (C) The structural comparison of T478K related to the Delta variant. (D) The structural demonstration of F490S related to the Lambda variant. (E) Demonstration of two mutants related to the two variants, L452R and L452Q of Delta and Lambda, respectively. (F) Demonstration of sequence deletion (RSYLTPG at position 246–252) and a mutation D253N located in the NTD of the spike protein of the Lambda variant.
The genomic data and protein sequence analysis of Delta and Lambda variants were compared with the original Wuhan isolate, available on the NCBI and GISAID clade database, accessed on 29 August 2021 from (http://gisaid.org). The comparison shows abundant changes in NSPs and S proteins (Table 1). The very large number of mutations in Delta and Lambda, combined with the limited experimental data, make the analysis of the significance of mutations extremely difficult at the current time. According to the available sequences of accessory proteins and comparison to the Wuhan strain, ORF6 and ORF7b of Delta and ORFs 6, 7a and 7b of Lambda and ORF10 of both variants had no changes. More information and amino acid sequences are available in Table A1. There are 20 substitutions in Delta’s NSP and 15 substitutions in Lambda’s NSP, plus 2 deletions of trimers. Within the S protein, Delta has 7 amino acid substitutions, while Lambda has 7 amino acid substitutions plus a heptad deletion. The Lambda changes provide replication advantages [52][31]. The substitutions stabilize the S protein with human ACE2, causing the virus to evade NAbs and increasing COVID-19 severity [6,48,52,53][26][27][31][32]. There is more conservation in E and M proteins. For Delta and Lambda, there are no changes in the E protein and single amino acid substitutions in the M protein. With regard to the N protein, Delta has 4 amino acid substitutions, while Lambda has 8.
Table 1. List of amino acid (aa) substitutions and deletions of Delta and Lambda variants in comparison with the original Wuhan strain. The non-structural proteins (NSPs) and accessory proteins in beige, envelope protein (E) in cyan, membrane protein (M) in green, nucleocapsid protein (N) in yellow, and spike glycoprotein (S) in dark orange color represent different changes for both Delta and Lambda variants of SARS-CoV-2, using GISAID ID: EPI_ISL_402124. The Lambda variant has more changes, compared to the Delta variant, and shows a deleted amino acid sequence in NSPs and S proteins. Delta and Lambda sequences extracted from NCBI tool and changes were found, using the GISAID online tool. Changes in accessory proteins ORF3, ORF7a and ORF8 in Delta, and ORF8 in Lambda are highlighted in dark blue color.
| COVID-19 Variant | aa Substitutions Deletions | Source | |||||
|---|---|---|---|---|---|---|---|
| NSPs Accessory Proteins | E | M | N | S | |||
| Delta (δ) | NSP3: | P822L, T1022X, P2767L, T2967X |
No Change | I82T | D63G, R203M, Q349H, D377Y | T19R, A222V, L452R, T478K, D614G, P681R, D950N |
NCBI, GISAID (EPI_ISL_402124) |
| NSP4: | A446V, A946V | ||||||
| NSP6: | V149A, T181I, V439A, T471I |
||||||
| NSP12: | P323L, M463I, G671S | ||||||
| NSP13: | P77L, T125N | ||||||
| ORF3: | S26L | ||||||
| ORF7a: | V82A, T120I | ||||||
| ORF8: | D119V, F120L | ||||||
| Lambda (λ) | NSP3: | T428I, P1469S, F1569V, T2373I, P3414S, F3514V |
No Change | K384N | P13L, R203K, G204R, G214C, P432L, R622K, G623R, G633C | G75V, T76I, R246Δ, S247Δ, Y248Δ, L249Δ, T250Δ, P251Δ, G252Δ, D253N, L452Q, F490S, D614G, T859N |
NCBI, GISAID (EPI_ISL_402124) |
| NSP4: | L438P, T492I, L938P, T992I |
||||||
| NSP5: | G15S, G321S | ||||||
| NSP6: | S106Δ, G107Δ, F108Δ, S396Δ, G397Δ, F398Δ |
||||||
| NSP12: | P323L | ||||||
| NSP13: | T599I | ||||||
| ORF8: | S142N | ||||||
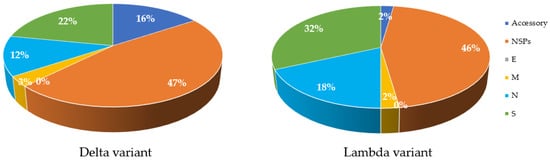
Mutations are a key factor to determine the characteristics of the virus in terms of severity and infectivity. The number of mutations in structural and non-structural proteins in both Delta and Lambda variants in comparison with the Wuhan isolate are available on GISAID (using CoVsurver: Mutation Analysis of hCoV-19), accessed on 29 August 2021, (https://www.gisaid.org/epiflu-applications/covsurver-mutations-app). In comparison to the original Wuhan strain, the Delta variant has 47% of its total differences in NSPs, while the Lambda variant has 46% (Figure 3). The Lambda variant also has more mutations in S and N proteins (Figure 2). In contrast, the M and E proteins of both variants have incurred one or no change (Figure 3). Analysis of the protein structure changes due to nucleotide sequence changes and determination of how these changes affect protein–protein interactions using bioinformatic tools can lead to new approaches for treatment. Proteins with high sequence conservation, such as E or M, may be a better choice for therapeutic purposes. Further details about the frequency of changes are listed in Table A2.


Figure 3. Comparison of amino acid changes in structural and Non-structural proteins in Delta and Lambda variants of SARS-CoV-2 compared to the Wuhan strain. Most changes are observed in NSP proteins (47% for Delta and 46% for Lambda), shown in orange colour in both variants. S proteins, shown in green color, have the greatest changes compared to other structural proteins. E protein, shown in grey color, has no mutations (0%) in both variants. Accessory proteins, shown in dark blue of Lambda show less change (2%) compared to Delta variant (16%). M proteins, shown in yellow color, have the least change in both variants, apart from E protein. The initial databases were extracted from GISAID online tool and processed using Excel.
References
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 256–258.
- Dhakal, S.; Macreadie, I.J.M.A. Genes of SARS-CoV-2 and emerging variants. Microbiol. Aust. 2021, 42, 10–12.
- Wang, H.; Li, X.; Li, T.; Zhang, S.; Wang, L.; Wu, X.; Liu, J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1629–1635.
- Sahin, E.; Bozdayi, G.; Yigit, S.; Hager, M.; Dizbay, M.; Tunccan, O.G.; Fidan, I.; Kayhan, C. Genomic characterization of SARS-CoV-2 isolates from patients in Turkey reveals the presence of novel mutations in spike and nsp12 proteins. J. Med. Virol. 2021, 93, 6016–6026.
- Cannalire, R.; Cerchia, C.; Beccari, A.R.; Di Leva, F.S.; Summa, V. Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities. J. Med. Chem. 2020.
- Rut, W.; Groborz, K.; Zhang, L.; Sun, X.; Zmudzinski, M.; Pawlik, B.; Wang, X.; Jochmans, D.; Neyts, J.; Młynarski, W.; et al. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021, 17, 222–228.
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821.
- da Silva, S.J.R.; Alves da Silva, C.T.; Mendes, R.P.G.; Pena, L. Role of nonstructural proteins in the pathogenesis of SARS-CoV-2. J. Med. Virol. 2020, 92, 1427–1429.
- Clark Lauren, K.; Green Todd, J.; Petit Chad, M.; Dutch Rebecca, E. Structure of Nonstructural Protein 1 from SARS-CoV-2. J. Virol. 2021, 95, e02019–e02020.
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216.
- Shang, J.; Han, N.; Chen, Z.; Peng, Y.; Li, L.; Zhou, H.; Ji, C.; Meng, J.; Jiang, T.; Wu, A. Compositional diversity and evolutionary pattern of coronavirus accessory proteins. Brief. Bioinform. 2021, 22, 1267–1278.
- Redondo, N.; Zaldivar-Lopez, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.J.T. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574.
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021, 54, 159–163.
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233.
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A Potential Therapeutic Target for COVID-19. iScience 2020, 23, 16–32.
- Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019, 5, 4580.
- Prompetchara, E.; Ketloy, C.; Tharakhet, K.; Kaewpang, P.; Buranapraditkun, S.; Techawiwattanaboon, T.; Sathean-anan-kun, S.; Pitakpolrat, P.; Watcharaplueksadee, S.; Phumiamorn, S.; et al. DNA vaccine candidate encoding SARS-CoV-2 spike proteins elicited potent humoral and Th1 cell-mediated immune responses in mice. PLoS ONE 2021, 16, e0248007.
- Mohammadi, M.; Shayestehpour, M.; Mirzaei, H. The impact of spike mutated variants of SARS-CoV2 on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021, 25, 101606.
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P.J.B. Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and its Receptor ACE2. bioRxiv 2021, 7, 19.
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant emergence and vaccine breakthrough. Nat. Portf. 2021.
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv 2021.
- Vu, M.N.; Menachery, V.D. Binding and Entering: COVID Finds a New Home. PLOS Pathog. 2021, 17, e1009857.
- Kimura, I.; Kosugi, Y.; Wu, J.; Yamasoba, D.; Butlertanaka, E.P.; Tanaka, Y.L.; Liu, Y.; Shirakawa, K.; Kazuma, Y.; Nomura, R.J.b. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. bioRxiv 2021, 10.
- Padilla-Rojas, C.; Jimenez-Vasquez, V.; Hurtado, V.; Mestanza, O.; Molina, I.S.; Barcena, L.; Morales Ruiz, S.; Acedo, S.; Lizarraga, W.; Bailon, H.; et al. Genomic analysis reveals a rapid spread and predominance of lambda (C.37) SARS-CoV-2 lineage in Peru despite circulation of variants of concern. J. Med. Virol. 2021.
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 7.
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280.
- Yan, R.; Zhang, Y.; Li, Y.; Ye, F.; Guo, Y.; Xia, L.; Zhong, X.; Chi, X.; Zhou, Q. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res. 2021, 31, 717–719.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H. Ferrin TEUCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82.
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224.
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383.
- Hristova, K.; Wimley, W.C. A look at arginine in membranes. J. Membr. Biol. 2011, 239, 49–56.
More
