Towards a comprehensive identification and functional characterization of nutrient-responsive ncRNAs and their downstream molecules, high-throughput sequencing has produced massive omics data for comparative expression profiling as a first step.
- microRNA
- long noncoding RNA
- circular RNA
- macronutrient
1. Introduction
Nutrient stress is one of the environmental adversities commonly encountered by plants. A thorough understanding of the adaptive strategy to various nutrient stresses will substantially strengthen the theoretical basis for plant breeding practices. Among 14 essential mineral nutrient elements for plant growth and development, six macronutrients, including nitrogen (N), phosphorus (P), potassium (K), calcium (Ca), magnesium (Mg), and sulfur (S), are required in relatively large amounts [1]. Although all these macronutrient elements are mainly absorbed from soil, they differ in sensing and transport pathways, as well as structural and metabolic functions, and may therefore induce different plant responses upon excessive or deficient nutrient supply.
N is the major component of amino acids, nucleotides, chlorophyll, vitamins, and alkaloids, with inorganic ammonium and nitrate being its main form for plant uptake [2]. Both insufficient and excessive levels of N can affect diverse aspects of the plant life cycle. For example, N deficiency could disturb the synthesis of chlorophyll and then reduce the level of photosynthesis, leading to decreased crop yield and quality [3], while an excessive application of N fertilizer might result in a decrease of sugar content [4]. Moderate N deficiency stimulates lateral root growth [5], and severe N deficiency inhibits lateral root growth [6]. P is another essential element involved in vital processes such as photosynthesis, respiration, signal transduction, and nucleic acid synthesis, and it can only be obtained from soil in the form of inorganic phosphorus (Pi) [7,8][7][8]. Pi deficiency could not only induce phenotypic changes including dark purple leaf and stem, reduced shoot, and more complex root growth [9], but also cause metabolite alterations such as reduced content of soluble sugar and increased content of organic acids and pigments [10]. K is the third most important macronutrient, and is primarily absorbed by plants in the form of K + . It mainly acts on plants via maintenance of cellular osmotic pressure, adjustment of enzyme activity, optimization of photosynthesis performance, and promotion of assimilation product transport [11]. The impacts of K deficiency on plant yield and quality have also been well documented in a variety of species [12,13,14,15,16,17,18][12][13][14][15][16][17][18]. Ca is a constituent of cell walls, and mainly participates in maintaining the cell physiological state in plants [19]. It is absorbed by plants in the form of Ca 2+ , a well-known second messenger in cellular signal transduction. A lack of Ca during the fruit ripening process might lead to leakage of cell membranes, irregular softening of cell walls, and abnormal fruit development [20]. Mg is involved in enzyme activation, cell homeostasis, membrane structure stability, active oxygen metabolism, nucleic acid metabolism, and signal transduction [21,22,23][21][22][23]. When Mg deficiency occurs in soil, plant growth is restricted, the leaf becomes yellow, and the biomass allocation between organs changes [24,25,26][24][25][26]. On the other hand, an excess of Mg could result in enhanced and weakened carbon metabolism in roots and leaves, respectively [27]. S is a component of amino acids, sulfated polysaccharides, sulfolipids, and vitamins [28], and plays a decisive role in the structure and biological activity of coenzymes and secondary metabolic products [29]. Plants mainly absorb S from soil in the form of inorganic sulfate through sulfate transporters. S deficiency can also lead to metabolite changes such as increased content of total phenol and reduced content of carotenoid in fruits [30,31][30][31].
Among the complex regulatory network underlying plant response to environmental stimuli, noncoding RNAs (ncRNAs) are a class of molecules that play critical roles in coordinating nutrient supply and plant demand. Currently, the most widely studied ncRNAs are microRNAs (miRNAs), which mainly act as negative regulators of their target genes through sequence-specific mRNA cleavage or translational repression [32,33][32][33]. In plants, miRNAs have long been known to participate in a wide range of biological processes indispensable for plant growth and stress responses [34,35][34][35]. Another subclass of ncRNAs that have been recently recognized as regulators for plant responses to biotic and abiotic stresses are long ncRNAs (lncRNAs) [36], which can either be processed into miRNAs to further modulate the expression of downstream genes, or function as molecular decoys to sequester small RNAs from their target RNAs [37]. In addition, there have also been sporadic reports on the potential involvements of other types of ncRNAs such as circular RNA (circRNA) and cis-natural antisense transcripts (cis-NATs) in nutrient stress response [38,39][38][39].
2. miRNA-Mediated Regulation in Response to Macronutrient Stress
Under nutrient stress, miRNA may be either upregulated or downregulated, and hence strengthen or relax its inhibition of target gene expression to adapt to variations in environmental nutrient concentrations. In most cases, the target genes of plant nutrient-responsive miRNAs may encode the sensor or transporter of a certain nutrient element, or the transcription factor in regulation of nutrient homeostasis. In this section, we will summarize major responsive proteins and genes for each macronutrient, with special interests in those targeted by nutrient-responsive miRNAs.
The phosphate starvation response (PHR) is a type of MYB transcription factor that regulates the expression of phosphate-starvation-induced (PSI) genes by binding to the P1BS motif (GNATATNC) in the promoter region. The expression of PHR1 is not sensitive to P starvation while regulated by the SPX-domain protein [60][40]. In a high P environment, AtSPX1 showed a high binding affinity to AtPHR1, which inhibited the binding of AtPHR1 to the P1BS motif of PSI genes; whereas under low P stress, the affinity between AtSPX1 and AtPHR1 was weakened, and AtPHR1 could activate the expression of PSI genes through binding to their P1BS motifs [61][41]. Among those PSI genes, the members of SPX-MFS subfamily, including phosphate transporter (PHT) proteins, are involved in intracellular P transport process. In case of P deficiency, the expression of PHT1 was directly induced by PHR1 to promote Pi uptake [62][42]. Nitrogen limitation adaptation (NLA), another SPX domain protein with E3 ubiquitin ligase activity, can coordinate with the E2 ligase PHO2 to modulate PHT1 degradation, while PHO2 also triggers degradation of PHO1 independent of NLA [63][43]. PHO1 is an SPX domain protein involved in xylem loading of Pi [64][44].
The main miRNAs involved in response to P stress are miR399 and miR827 (Figure 1). In Arabidopsis, Ath-miR399 was specifically induced by low P, and it could recognize the AtPHO2 gene and thereby regulate P homeostasis and signaling pathways in plants [65,66,67][45][46][47]. Currently, miR399 has been found in rice, tomato, alfalfa, kidney bean, and strawberry, showing induced expression by P deficiency stress [66,67,68,69][46][47][48][49]. At the same time, inhibited expression of PHO2 homologous gene by miR399 was also found in rice and kidney bean under low P conditions [70,71][50][51]. Interestingly, long-distance movement of miR399s from shoots to roots was discovered in Arabidopsis, and was suggested to be crucial for enhancing Pi uptake and translocation during the onset of Pi deficiency [72][52]. Later, miR399 and miR395 were also observed to be phloem-mobile in Brassica under nutrient starvation [73][53]. The function of miR827 was not conserved among different species [74][54]. Although miR827 was strongly induced under P stress in Arabidopsis and rice, the target gene of Ath-miR827 was AtNLA , while the target of Osa-miR827 was OsPHT5 [75,76][55][56]. Similar to miR399, translocation of miR827 and miR2111a between shoots and roots during Pi starvation was also evident in Arabidopsis [77][57]. In addition, Ath-miR156 and its target, the AtSPL ( squamosa promoter binding protein-like ) gene, were also induced by P stress and participated in stress response by regulating the accumulation of organic acids and anthocyanins in the rhizosphere [78][58].
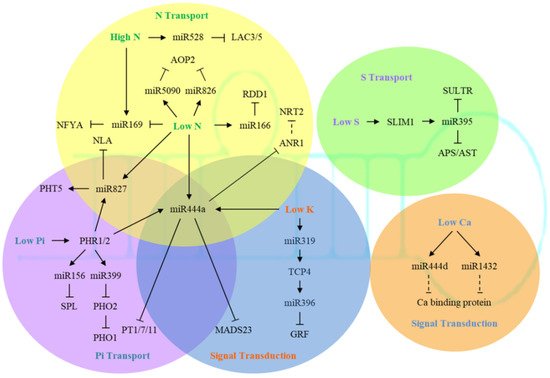
Several studies have identified a large number of K + -transporting proteins in multiple gene families [79][59]. Among them, AtHAK5 from the KT/KUP/HAK family may function in both low-affinity and high-affinity transport, and is strongly induced by K deficiency [80,81,82,83][60][61][62][63]. The expression of AtHAK5 was regulated by AP2/ERF transcription factor AtRAP2.11, and could also modulate plant response to low K conditions [84][64]. So far, there have been 71 K + transporters and channel proteins identified in Arabidopsis. These proteins are not only involved in K uptake and transport in tissues and organs, but also are associated with K storage in vacuoles [85,86,87,88,89][65][66][67][68][69].
3. Large-Scale Identification of miRNAs Responsive to Differential Nutrient Availability
Systematic identification of candidate miRNAs involved in plant response to macronutrient stress has been performed in increasing number of species through comparative expression profiling of miRNAs among differentially adapted genotypes or the same genotype under differential nutrient availability, producing large amounts of small RNA sequencing (sRNA-Seq) data (Table 1). Global screening of miRNAs responsive to N starvation was first reported in Arabidopsis, in which miR160, miR780, miR826, miR842, and miR846 exhibited increased expression, while miR169, miR171, miR395, miR397, miR398, miR399, miR408, miR827, and miR857 showed decreased expression upon N stress [57][70]. Likewise, comparative expression profiling by deep sequencing helped to identified N-responsive miRNAs from shoots and roots of 7-day N-starved rice [50][71], from leaves and roots of two wheat cultivars subjected to chronic or short-term N stress [109,110][72][73], and from shoots and roots of rapeseeds with 0 or 72 h of N-limitation treatment [111][74].
Table 1. Summary for large-scale identification of macronutrient-stress-responsive miRNAs via high-throughput sequencing.
| Macronutrient Status | Data Quantity | Differentially Expressed miRNAs * | Species/Genotype ** | Tissue | Reference |
|---|---|---|---|---|---|
| N deficiency | 368.1 Mb | 9 ↓, 5 ↑ | Arabidopsis thaliana/Columbia | Seedling | [57][70] |
| 93.8 Mb | 9 ↓, 13 ↑ | Arabidopsis thaliana/Columbia | Seedling | [112][75] | |
| 26.6 Gb | 5 ↓, 30 ↑ | Oryza sativa/Nipponbare | Shoot | [50][71] | |
| 57 ↓, 15 ↑ | Root | ||||
| 1.5 Gb | 2 ↓, 1 ↑ | Triticum turgidum/Svevo | Flag Leaf & Spike | [109,110][72][73] | |
| 4 ↓, 5 ↑ | Leaf & Stem | ||||
| 6 ↓, 5 ↑ | Root | ||||
| 1.5 Gb | 2 ↓, 3 ↑ | Triticum turgidum/Ciccio | Flag Leaf & Spike | ||
| 4 ↓ | Leaf & Stem | ||||
| 3 ↓, 4 ↑ | Root | ||||
| 3.4 Gb | 71 ↓, 52 ↑ | Brassica napus /Zhongshuang11 | Shoot | [111][74] | |
| 64 ↓, 37 ↑ | Root | ||||
| 3.8 Gb | 28 ↓, 8 ↑ | Sorghum bicolor/BTX623 | Shoot | [113][76] | |
| 25 ↓, 13 ↑ | Root | ||||
| P deficiency | 132.6 Mb | 21 ↑ | Arabidopsis thaliana/Columbia | Shoot | [53][77] |
| 657.6 Mb | 22 ↓, 33 ↑ | Arabidopsis thaliana/Columbia | Shoot | [75][55] | |
| 20 ↓, 25 ↑ | Root | ||||
| 961.9 Mb | 27 ↓, 7 ↑ | Glycine max/BX10 | Root | [114][78] | |
| 40 ↓, 12 ↑ | Shoot | ||||
| 4.3 Gb | 24 ↓, 22 ↑ | Glycine max/Bogao | Root | [115][79] | |
| 4.4 Gb | 49 ↓, 34 ↑ | Glycine max/Nannong94-156 | Root | ||
| 27.6 Mb | 3 ↓, 2 ↑ | Zea mays/Inbred line 178 | Root | [116][80] | |
| 14.2 Gb | 174 | Zea mays/Inbred line Q319 | Leaf & Root | [117][81] | |
| 4.2 Gb | 16 ↓, 33↑ | Sorghum bicolor/BTX623 | Shoot | [113][76] | |
| 58 ↓, 18↑ | Root | ||||
| K deficiency | 4.4 Gb | 22 ↓, 25 ↑ | Hordeum vulgare/XZ149 | Seedling | [91][82] |
| 4.5 Gb | 21 ↓, 17 ↑ | Hordeum vulgare/ZD9 | Seedling | ||
| 4.2 Gb | 7 ↓, 5 ↑ | Triticum aestivum/Kenong9204 | Root | [118][83] | |
| 3.2 Gb | 110 ↓, 122 ↑ | Solanum lycopersicum/JZ18 vs. 35S:SlmiR168a | Leaflet | [119][84] | |
| 3.8 Gb | 58 ↓, 102 ↑ | Solanum lycopersicum/JZ18 vs. 35S:rSlAGO1 | Leaflet | ||
| 4.0 Gb | 12 ↓, 20 ↑ | Sorghum bicolor/BTX623 | Shoot | [113][76] | |
| 16 ↓, 6 ↑ | Root | ||||
| Mg deficiency | 2.0 Gb | 71 ↓, 75 ↑ | Citrus sinensis/Xuegan | Leaf | [107][85] |
| 1.0 Gb | 69 ↓, 101 ↑ | Citrus sinensis/Xuegan | Root | [108][86] | |
| Ca deficiency | 7.0 Gb | 87 | Arachis hypogea/Baisha1016 | Embryo | [120][87] |
| S deficiency | 101.9 Mb | 2 ↓, 2 ↑ | Arabidopsis thaliana/Columbia | Seedling | [112][75] |
The early attempts at large-scale identification of potential P-responsive miRNAs were also reported in Arabidopsis (Table 1), in which the expression of miR156, miR399, miR778, miR827, and miR2111 was induced, but the expression of miR169, miR395, and miR398 was repressed upon P deprivation [53,75][77][55]. Since then, an increasing number of candidate P-responsive miRNAs have been obtained in other plant species using different strategies. A microarray-based approach successfully uncovered a subset of 57 known plant miRNAs with differential expression in leaves or roots of soybeans grown under P-deficient and P-sufficient conditions [121][88], while a genomewide mining dependent on sRNA-Seq identified not only conserved, but also novel miRNAs with significantly altered expression in roots or shoots of a P-efficient genotype soybean treated with low P and high P [114][78]. Recently, 777 differentially expressed miRNAs across different P treatments and soybean genotypes were also screened out by deep sequencing [115][79]. Not surprisingly, the sequencing-based expression profiling resulted in substantially larger number of candidate miRNAs than the array-based method did for the same species. In addition, systematic screening of P-responsive miRNAs was also achieved in major crops including rice [122][89], maize [116[80][81],117], and wheat [123][90].
Global identification of K-deficiency-responsive miRNAs was conducted in roots of two barley genotypes differing in low K tolerance, as well as in wheat roots under five periods of low K treatments, generating approximately 9 Gb and 4 Gb of sRNA-Seq data, respectively ( Table 1 ). The former detected 28 miRNAs differentially expressed at both 2 days and 10 days after low K stress for two barley genotypes [91][82], while the latter found miR9772, miR1120b-3p, miR531, and miR319 displaying differential expression at all time points during the low K treatments, and suggested that these miRNAs were most possibly involved in mediating plant adaptation to K deficiency [118][83]. Interestingly, high-throughput sequencing was also employed for identifying differentially expressed miRNAs between two transgenic tomato plants, separately overexpressing SlmiR168a and SlAGO1 , to explore downstream miRNAs (miR171, miR384, miR530, miR858, and miR8007) involved in the SlmiR168 -mediated SlAGO1A regulation upon K stress [119][84].
Although the basic principle for identifying nutrient-responsive miRNA was to single out candidates with significantly differential expression between or among samples, the calculation method for expression level and the criteria for statistical significance varied among studies. Furthermore, the tissues for sampling also differed among studies, with roots being most frequently used, owing to their high susceptibility to variations in environmental nutrient levels ( Table 1 ). In contrast, there was only one work focusing on the influence of nutrient stress on miRNA abundance in the reproductive tissue of peanut [120][87]. The lack of progress in achieving nutrient-responsive miRNAs in reproductive tissues might also be attributed to the fact that most of the nutrient-stressed conditions applied in these studies would cause severe symptoms in vegetative tissues and result in failure of flowering or fruit setting. Nevertheless, the results from vegetative tissues still suggested tissue-specific miRNA regulation upon the same type of macronutrient stress. For instance, 13 miRNAs showed similar expression changes in roots and shoots of soybeans under P deficiency, while 6 miRNAs had opposite expression changes in these two tissues [114][78]. In rapeseed, 11 upregulated and 15 downregulated miRNAs were specifically identified in roots under N starvation, whereas 25 upregulated and 23 downregulated miRNAs were specifically identified in shoots [111][74].
4. Other Types of ncRNAs Involved in Nutrient Stress Response
5. Conclusions and Perspectives
References
- Nath, M.; Tuteja, N. NPKS uptake, sensing, and signaling and miRNAs in plant nutrient stress. Protoplasma 2016, 253, 767–786.
- Hachiya, T.; Sakakibara, H. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 2017, 68, 2501–2512.
- Wen, B.; Li, C.; Fu, X.; Li, D.; Li, L.; Chen, X.; Wu, H.; Cui, X.; Zhang, X.; Shen, H.; et al. Effects of nitrate deficiency on nitrate assimilation and chlorophyll synthesis of detached apple leaves. Plant Physiol. Biochem. 2019, 142, 363–371.
- Smoleń, S.; Sady, W. The effect of various nitrogen fertilization and foliar nutrition regimes on the concentrations of sugars, carotenoids and phenolic compounds in carrot (Daucus carota L.). Sci. Hort. 2009, 120, 315–324.
- Gruber, B.D.; Giehl, R.F.; Friedel, S.; von Wiren, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179.
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wiren, N.; Takahashi, H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034.
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206.
- Baker, A.; Ceasar, S.A.; Palmer, A.J.; Paterson, J.B.; Qi, W.; Muench, S.P.; Baldwin, S.A. Replace, reuse, recycle: Improving the sustainable use of phosphorus by plants. J. Exp. Bot. 2015, 66, 3523–3540.
- Zhang, Y.; Zhou, Y.; Chen, S.; Liu, J.; Fan, K.; Li, Z.; Liu, Z.; Lin, W. Gibberellins play dual roles in response to phosphate starvation of tomato seedlings, negatively in shoots but positively in roots. J. Plant Physiol. 2019, 234–235, 145–153.
- Li, Z.; Qiu, Q.; Chen, Y.; Lin, D.; Huang, J.; Huang, T. Metabolite alteration in response to low phosphorus stress in developing tomato fruits. Plant Physiol. Biochem. 2021, 159, 234–243.
- Li, W.; Xu, G.; Alli, A.; Yu, L. Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 2018, 74, 133–141.
- Preciado-Rangel, P.; Troyo-Dieguez, E.; Valdez-Aguilar, L.A.; Garcia-Hernandez, J.L.; Luna-Ortega, J.G. Interactive effects of the potassium and nitrogen relationship on yield and quality of strawberry grown under soilless conditions. Plants 2020, 9, 441.
- Lester, G.E.; Jifon, J.L.; Makus, D.J. Supplemental foliar potassium applications with or without a surfactant can enhance netted muskmelon quality. Hort. Sci. 2006, 41, 741–744.
- Kumar, D.; Ahmed, N. Response of nitrogen and potassium fertigation to “Waris” almond (Prunus dulcis) under northwestern Himalayan Region of India. Sci. World J. 2014, 2014, 141328.
- Wang, Y.; Lv, J.; Chen, D.; Zhang, J.; Qi, K.; Cheng, R.; Zhang, H.; Zhang, S. Genome-wide identification, evolution, and expression analysis of the KT/HAK/KUP family in pear. Genome 2018, 61, 755–765.
- Zhang, W.; Zhang, X.; Wang, Y.; Zhang, N.; Guo, Y.; Ren, X.; Zhao, Z. Potassium fertilization arrests malate accumulation and alters soluble sugar metabolism in apple fruit. Biol. Open 2018, 7, bio024745.
- Shen, C.; Shi, X.; Xie, C.; Li, Y.; Yang, H.; Mei, X.; Xu, Y.; Dong, C. The change in microstructure of petioles and peduncles and transporter gene expression by potassium influences the distribution of nutrients and sugars in pear leaves and fruit. J. Plant Physiol. 2019, 232, 320–333.
- Yener, H.; Altuntaş, Ö. Effects of potassium fertilization on leaf nutrient content and quality attributes of sweet cherry fruits (Prunus avium L.). J. Plant Nutr. 2020, 44, 946–957.
- Marschner, P. Marscher’s Mineral Nutrition of High Plants, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 135–189.
- Michailidis, M.; Karagiannis, E.; Tanou, G.; Sarrou, E.; Stavridou, E.; Ganopoulos, I.; Karamanoli, K.; Madesis, P.; Martens, S.; Molassiotis, A. An integrated metabolomic and gene expression analysis identifies heat and calcium metabolic networks underlying postharvest sweet cherry fruit senescence. Planta 2019, 250, 2009–2022.
- Gerendás, J.; Führs, H. The significance of magnesium for crop quality. Plant Soil 2013, 368, 101–128.
- Guo, W.; Chen, S.; Hussain, N.; Cong, Y.; Liang, Z.; Chen, K. Magnesium stress signaling in plant: Just a beginning. Plant Signal. Behav. 2015, 10, e992287.
- Wang, Z.; Hassan, M.U.; Nadeem, F.; Wu, L.; Zhang, F.; Li, X. Magnesium fertilization improves crop yield in most production systems: A meta-analysis. Front. Plant Sci. 2019, 10, 1727.
- Hermans, C.; Verbruggen, N. Physiological characterization of Mg deficiency in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 2153–2161.
- Saghaiesh, S.P.; Souri, M.K.; Moghaddam, M. Effects of different magnesium levels on some morphophysiological characteristics and nutrient elements uptake in Khatouni melons (Cucumis melo var. inodorus). J. Plant Nutr. 2018, 42, 27–39.
- Ding, Y.; Luo, W.; Xu, G. Characterisation of magnesium nutrition and interaction of magnesium and potassium in rice. Ann. App. Biol. 2006, 149, 111–123.
- Kwon, M.C.; Kim, Y.X.; Lee, S.; Jung, E.S.; Singh, D.; Sung, J.; Lee, C.H. Comparative metabolomics unravel the effect of magnesium oversupply on tomato fruit quality and associated plant metabolism. Metabolites 2019, 9, 231.
- Leustek, T.; Martin, M.N.; Bick, J.; Davies, J.P. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 141–165.
- Davidian, J.C.; Kopriva, S. Regulation of sulfate uptake and assimilation-the same or not the same? Mol. Plant 2010, 3, 314–325.
- Thomas, S.G.; Bilsborrow, P.E.; Hocking, T.J.; Bennett, J. Effect of sulphur deficiency on the growth and metabolism of sugar beet (Beta vulgaris cv Druid). J. Sci. Food Agri. 2000, 80, 2057–2062.
- Mohammed, K.A.S.; Hellal, F.A.; EL-Sayed, S.A.A. Influence of sulfur deprivation on biomass allocation, mineral composition and fruit quality of tomato plants. Mid. East J. Agri. Res. 2015, 4, 42–48.
- Bartel, D.P. MicroRNAs: Genomics biogenesis, mechanism, and function. Cell 2004, 11, 281–297.
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114.
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulation roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53.
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203.
- Nejat, N.; Mantri, N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol. 2018, 38, 93–105.
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407.
- Wang, W.; Wang, J.; Wei, Q.; Li, B.; Zhong, X.; Hu, T.; Hu, H.; Bao, C. Transcriptome-wide identification and characterization of circular RNAs in leaves of Chinese cabbage (Brassica rapa L. ssp. pekinensis) in response to calcium deficiency-induced tip-burn. Sci. Rep. 2019, 9, 14544.
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genom. Proteom. Bioinf. 2015, 13, 137–147.
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133.
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; de Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodriguez, J.; et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952.
- Liu, J.; Yang, L.; Luan, M.; Wang, Y.; Zhang, C.; Zhang, B.; Shi, J.; Zhao, F.G.; Lan, W.; Luan, S. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 6571–6578.
- Yue, W.; Ying, Y.; Wang, C.; Zhao, Y.; Dong, C.; Whelan, J.; Shou, H. OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 2017, 90, 1040–1051.
- Cao, F.; Li, H.; Wang, S.; Li, X.; Dai, H.; Zhang, Z. Expression and functional analysis of FaPH01;H9 gene of strawberry (Fragaria x ananassa). J. Integr. Agr. 2017, 16, 580–590.
- Fujii, H.; Chiou, T.J.; Lin, S.I.; Aung, K.; Zhu, J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043.
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999.
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421.
- Branscheid, A.; Sieh, D.; Pant, B.D.; May, P.; De Vers, E.A.; Elkrog, A.; Schauser, L.; Scheible, W.R.; Krajinski, F. Expression pattern suggests a role of miR399 in the regulation of the cellular response to local Pi increase during arbuscular mycorrhizal symbiosis. Mol. Plant-Microbe Interact. 2010, 23, 915–926.
- Liu, J.Q.; Allan, D.L.; Vance, C.P. Systemic signaling and local sensing of phosphate in common bean: Cross-talk between photosynthate and microRNA399. Mol. Plant 2010, 3, 428–437.
- Valdes-Lopez, O.; Arenas-Huertero, C.; Ramirez, M.; Girard, L.; Sanchez, F.; Vance, C.P.; Luis Reyes, J.; Hernandez, G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008, 31, 1834–1843.
- Hu, B.; Wang, W.; Deng, K.; Li, H.; Zhang, Z.; Zhang, L.; Chu, C. MicroRNA399 is involved in multiple nutrient starvation responses in rice. Front. Plant. Sci. 2015, 6, 188.
- Lin, S.I.; Chiang, S.; Lin, W.; Chen, J.; Tseng, C.; Wu, P.; Chiou, T.J. Regulation network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 2008, 147, 723–746.
- Buhtz, A.; Pieritz, J.; Springer, F.; Kehr, J. Phloem small RNAs, nutrient stress reponses, and systemic mobility. BMC Plant Biol. 2010, 10, 64.
- Lin, W.; Lin, Y.; Chiang, S.F.; Syu, C.; Hsieh, L.C.; Chiou, T.J. Evolution of microRNA827 targeting in the plant kingdom. New Phytol. 2018, 217, 1712–1725.
- Hsieh, L.; Lin, S.I.; Shih, A.C.; Chen, J.; Lin, W.; Tseng, C.; Li, W.; Chiou, T. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132.
- Lin, S.; Santi, C.; Jobet, E.; Lacut, E.; El Kholti, N.; Karlowski, W.M.; Verdeil, J.L.; Breitler, J.C.; Perin, C.; Ko, S.S.; et al. Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 2010, 51, 2119–2131.
- Huen, A.K.; Rodriguez-Medina, C.; Ho, A.Y.Y.; Atkins, C.A.; Smith, P.M.C. Long-distance movement of phosphate starvation-responsive microRNAs in Arabidopsis. Plant Biol. 2017, 19, 643–649.
- Lei, K.J.; Lin, Y.M.; An, G.Y. miR156 modulates rhizosphere acidification in response to phosphate limitation in Arabidopsis. J. Plant Res. 2016, 129, 275–284.
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.M.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667.
- Ahn, S.J.; Shin, R.; Schachtman, D.P. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 2004, 134, 1135–1145.
- Gierth, M.; Maser, P.; Schroeder, J.I. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137, 1105–1114.
- Qi, Z.; Hampton, C.R.; Shin, R.; Barkla, B.J.; White, P.J.; Schachtman, D.P. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 2008, 59, 595–607.
- Luan, S.; Lan, W.; Chul Lee, S. Potassium nutrition, sodium toxicity, and calcium signaling: Connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 2009, 12, 339–346.
- Kim, M.J.; Ruzicka, D.; Shin, R.; Schachtman, D.P. The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant 2012, 5, 1042–1057.
- Wang, Y.; Wu, W.H. Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 2017, 39, 123–128.
- Wang, Y.; Chen, Y.F.; Wu, W.H. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2021, 63, 34–52.
- Gierth, M.; Maser, P. Potassium transporters in plants-involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 2007, 581, 2348–2356.
- Chanroj, S.; Wang, G.; Venema, K.; Zhang, M.W.; Delwiche, C.F.; Sze, H. Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 2012, 3, 25.
- Gomez-Porras, J.L.; Riano-Pachon, D.M.; Benito, B.; Haro, R.; Sklodowski, K.; Rodriguez-Navarro, A.; Dreyer, I. Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 2012, 3, 167.
- Liang, G.; He, H.; Yu, D. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951.
- Shin, S.Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.K.; Shin, C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genom. 2018, 19, 532.
- Zuluaga, D.L.; Domenico, D.P.; Michela, J.; Luca, C.P.; Gabriella, S.; Turgay, U. Durum wheat miRNAs in response to nitrogen starvation at the grain filling stage. PLoS ONE 2017, 12, e0183253.
- Zuluaga, D.L.; Vittoria, L.; Luca, C.P.; Gabriella, S. MicroRNAs in durum wheat seedlings under chronic and short-term nitrogen stress. Funct. Integr. Genom. 2018, 18, 645–657.
- Hua, Y.; Zhou, T.; Huang, J.; Yue, C.; Song, H.; Guan, C.; Zhang, Z. Genome-wide differential DNA methylation and miRNA expression profiling reveals epigenetic regulatory mechanisms underlying nitrogen-limitation-triggered adaptation and use efficiency enhancement in allotetraploid rapeseed. Int. J. Mol. Sci. 2020, 21, 8453.
- Liang, G.; Ai, Q.; Yu, D. Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci. Rep. 2015, 5, 11813.
- Zhu, Z.; Li, D.; Cong, L.; Lu, X. Identification of microRNAs involved in crosstalk between nitrogen, phosphorus and potassium under multiple nutrient deficiency in sorghum. Crop J. 2021, 9, 465–475.
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.R. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009, 150, 1541–1555.
- Sha, A.; Chen, Y.; Ba, H.; Shan, Z.; Zhang, X.; Wu, X.; Qiu, D.; Chen, S.; Zhou, X. Identification of Glycine max microRNAs in response to phosphorus deficiency. J. Plant Biol. 2012, 55, 268–280.
- Liu, X.; Chu, S.; Sun, C.; Xu, H.; Zhang, J.; Jiao, Y.; Zhang, D. Genome-wide identification of low phosphorus responsive microRNAs in two soybean genotypes by high-throughput sequencing. Funct. Integr. Genom. 2020, 20, 825–838.
- Zhang, Z.; Lin, H.; Shen, Y.; Gao, J.; Xiang, K.; Liu, L.; Ding, H.; Yuan, G.; Lan, H.; Zhou, S.; et al. Cloning and characterization of miRNAs from maize seedling roots under low phosphorus stress. Mol. Biol. Rep. 2012, 39, 8137–8146.
- Gupta, S.; Kumari, M.; Kumar, H.; Varadwaj, P.K. Genome-wide analysis of miRNAs and Tasi-RNAs in Zea mays in response to phosphate deficiency. Funct. Integr. Genom. 2017, 17, 335–351.
- Zeng, J.; Ye, Z.; He, X.; Zhang, G. Identification of microRNAs and their targets responding to low-potassium stress in two barley genotypes differing in low-K tolerance. J. Plant Physiol. 2019, 234–235, 44–53.
- Zhao, Y.; Xu, K.; Liu, G.; Li, S.; Zhao, S.; Liu, X.; Yang, X.; Xiao, K. Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 15812.
- Liu, X.; Tan, C.; Cheng, X.; Zhao, X.; Li, T.; Jiang, J. miR168 targets Argonaute1A mediated miRNAs regulation pathways in response to potassium deficiency stress in tomato. BMC Plant Biol. 2020, 20, 447.
- Ma, C.L.; Qi, Y.P.; Liang, W.W.; Yang, L.T.; Lu, Y.B.; Guo, P.; Ye, X.; Chen, L.S. MicroRNA regulatory mechanisms on citrus sinensis leaves to magnesium-deficiency. Front. Plant Sci. 2016, 7, 201.
- Liang, W.; Huang, J.; Li, C.; Yang, L.; Ye, X.; Lin, D.; Chen, L. MicroRNA-mediated responses to long-term magnesium-deficiency in Citrus sinensis roots revealed by Illumina sequencing. BMC Genom. 2017, 18, 657.
- Chen, H.; Yang, Q.; Chen, K.; Zhao, S.; Zhang, C.; Pan, R.; Cai, T.; Deng, Y.; Wang, X.; Chen, Y.; et al. Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 392.
- Zeng, H.Q.; Zhu, Y.Y.; Huang, S.Q.; Yang, Z.M. Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J. Plant Physiol. 2010, 167, 1289–1297.
- Secco, D.; Whelan, J. Toward deciphering the genome-wide transcriptional responses of rice to phosphate starvation and recovery. Plant Signal. Behav. 2014, 9, 4285–4304.
- Zhao, X.; Liu, X.; Guo, C.; Gu, J.; Xiao, K. Identification and characterization of microRNAs from wheat (Triticum aestivum L.) under phosphorus deprivation. J. Plant Biochem. Biotechnol. 2012, 22, 113–123.
- Jabnoune, M.; Secco, D.; Lecampion, C.; Robaglia, C.; Shu, Q.; Poirier, Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 2013, 25, 4166–4182.
- Liu, F.; Xu, Y.; Chang, K.; Li, S.; Liu, Z.; Qi, S.; Jia, J.; Zhang, M.; Crawford, N.M.; Wang, Y. The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. New Phytol. 2019, 224, 117–131.
- Chen, M.; Wang, C.; Bao, H.; Chen, H.; Wang, Y. Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genom. 2016, 291, 1663–1680.
- Lv, Y.; Liang, Z.; Ge, M.; Qi, W.; Zhang, T.; Lin, F.; Peng, Z.; Zhao, H. Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genom. 2016, 17, 350.
- Chen, Z.; Jiang, Q.; Jiang, P.; Zhang, W.; Huang, J.; Liu, C.; Halford, N.G.; Lu, R. Novel low-nitrogen stress-responsive long non-coding RNAs (lncRNA) in barley landrace B968 (Liuzhutouzidamai) at seedling stage. BMC Plant Biol. 2020, 20, 142.
- Wang, J.; Chen, Q.; Wu, W.; Chen, Y.; Zhou, Y.; Guo, G.; Chen, M. Genome-wide analysis of long non-coding RNAs responsive to multiple nutrient stresses in Arabidopsis thaliana. Funct. Integr. Genom. 2021, 21, 17–30.
- Fukuda, M.; Nishida, S.; Kakei, Y.; Shimada, Y.; Fujiwara, T. Genome-wide analysis of long intergenic noncoding RNAs responding to low-nutrient conditions in Arabidopsis thaliana: Possible involvement of Trans-Acting siRNA3 in response to low nitrogen. Plant Cell Physiol. 2019, 60, 1961–1973.
- Li, F.; Shi, T.; He, A.; Huang, X.; Gong, J.; Yi, Y.; Zhang, J. Bacillus amyloliquefaciens LZ04 improves the resistance of Arabidopsis thaliana to high calcium stress and the potential role of lncRNA-miRNA-mRNA regulatory network in the resistance. Plant Physiol. Biochem. 2020, 151, 166–180.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Lv, L.; Yu, K.; Lu, H.; Zhang, X.; Liu, X.; Sun, C.; Xu, H.; Zhang, J.; He, X.; Zhang, D. Transcriptome-wide identification of novel circular RNAs in soybean in response to low-phosphorus stress. PLoS ONE 2020, 15, e0227243.
- He, H.; Liang, G.; Li, Y.; Wang, F.; Yu, D. Two young microRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol. 2014, 164, 853–865.
- Zhao, Q.; Zeng, D.; Fan, Z.; Yu, Z.; Hu, Y.; Zhang, J. Seasonal variations in phosphorus fractions in semiarid sandy soils under different vegetation types. For. Ecol. Manag. 2009, 258, 1376–1382.
