You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Koichi Takaki.
Pulsed power is the technology of accumulating energy during a relatively long period of time and of releasing the accumulated energy in an extremely short period of time as a high-power pulse composed of a high voltage and a large current but moderately low energy. The food processing applications of pulsed power technologies, such as intense pulsed electric fields and time-modulated discharge plasmas, have been studied worldwide. Pulsed electric fields (PEFs), which are generated by pulsed power technologies, are being tested for their applicability in food processing through protein conformational change and the poration of cell membranes.
- pulse electric field
- protein conformational change activity
1. Protein Conformational Change by PEF Irradiation
Intense PEFs are being tested for their potential uses in food processing, such as the inactivation of bacteria, enzyme activity control and fermentation acceleration, as non-thermal processing can have a reduced influence on food quality [1,9][1][2]. The advantage of PEF treatment compared to a thermal process is that it reduces detrimental changes in nutrition by retaining the physical and sensorial qualities of food [27][3]. PEF treatment is applied to a wide range of foods such as liquid (juice, milk, beer, etc.), semi-solid (gel-state foods) and solid-state foods. The typical operation range of PEF treatments is an electric field amplitude of 5–50 kV/cm with a pulse length in the range of several to tens of μs. PEFs are applied to food located between two electrodes and causes bacteria and enzymatic inactivation at a temperature lower than in thermal treatment [8,28][4][5]. The enzymatic reactions are a basic function of proteins, which are determined by its conformation of the polypeptide chain. Therefore, enzyme activity is affected by protein conformational change, such as the misfolding of proteins. The protein conformation consists of secondary structures (such as α-helices and β-sheets) and tertiary structures. The exposure of proteins to intense PEFs causes a conformational change through processes of electrical charging-up and the displacement of elements by electrical force [29][6]. In this section, the protein conformational change and enzyme inactivation by PEF irradiation are reviewed.
2. Conformational Change in Proteins
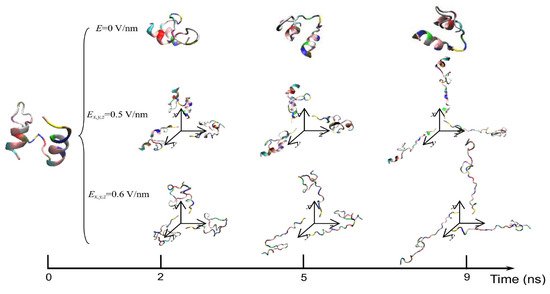
The exposure of proteins to intense PEFs causes conformational structural change through directly or indirectly affecting the secondary and tertiary structures. The direct effect of intense PEFs is a stretching of the molecular bindings in a protein, i.e., an unfolding structure caused by an electrostatic tensile force. Jiang et al. reported that the secondary structure was predicted to change from a helix into turns or random coils at an electric field with a strength of E > 0.5 V/nm (5.0 × 108 V/m) using molecular dynamics (MD) calculations as shown in Figure 15 [30][7]. The MD calculations are carried out for the 1BBL (consisting of 37 amino acid residues) protein molecule that includes two α-helix secondary fragments. The original 1BBL protein structure still remains when there is no exposure to an electric field for all orientations during the period set in the calculation. However, the protein is stretched for all orientations with exposure to an electric field with relatively high strength. This result indicates that the realignment of some charged residues is induced by the exposure to an electric field. The structure change from an α-helix to the turns and random coils is caused to be more rapid by increasing the electric field strength.
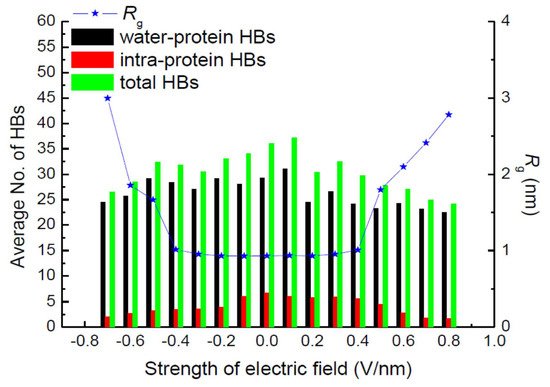
Hydrogen bonds (HBs) are important in their role of stabilizing the conformation of, e.g., secondary structures. Jiang et al. also calculated the average number of total HBs in protein structures using MD simulation at various strengths of an exposed electric field, as shown in Figure 162 [30][7]. The average number of intra-protein HBs decreases with an increasing electric field strength larger than 0.5 V/nm. The number of intra-protein HBs has a strong relationship with the conformational structure stability of the protein. Moreover, the radius of gyration, Rg, has the opposite tendency, against that of the number of intra-protein HBs.
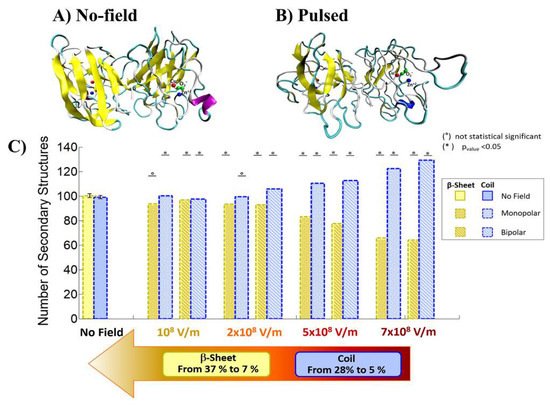
Qin and Buehler reported that the protein secondary structural transitions depended on the amino acid chain length. The short amino chain proteins with fewer than 26 amino acids (i.e., 3.8 nm in length) are easily induced as interprotein sliding. However, the long amino chain proteins with larger length causes a conformational change from α-helix to β-sheet, which lead to increase the protein stiffness, strength, and energy dissipation capacity [31,32][8][9]. Valle et al. reported MD analysis of the conformational change of a single superoxide dismutase (SOD1) enzyme by exposing it to a 100-ns-wide intense PEF in the range of 108 to 7 × 108 V/m in strength [33,34][10][11]. In the MD calculations, a monopolar (MP) or a bipolar (BP) 100 ns PEF is applied to SOD1. The intensity of 7 × 108 V/m induces a dramatic structural change with an irreversible transition from β-sheets or coil structures to unfolded states, as shown in Figure 173 [33][10].
Ding et al. calculated the electric force on the proteins which induces the conformational change with applied forces relative to the inter-chain bonding forces [35][12]. The inter-chain bonding of HBs in the α-helix and β-sheet was 8.1 kJ/mol (1.93 kcal/mol) and 6.6 kJ/mol (1.58 kcal/mol), respectively. Using the bonding energies of HBs and a distance between the elements of 0.35 nm, the inter-chain bonding forces of HB are obtained as 40 pN, which corresponds to approximately 108 V/m in electric field strength. The transition in conformational structure from α-helices to β-structures was also analyzed based on the four-bead model using discrete MD modeling. The potential energy (εHB) of a β-hairpin structure is larger than that of an α-helix. However, the entropy of a β-hairpin is larger than that of an α-helix. From the free energy of the HB for α-helix and β-hairpin conformations, the α-helix-to-β-hairpin transition is predicted to be caused at 0.125 εHB of the temperature. Here, the connections of primary structures consist of covalent bonds such as peptide bonds and disulfide bonds (S–S). These bonds have almost one order higher bonding energy (210–630 kJ/mol). For this reason, the primary structure is generally less sensitive to electric fields compared to secondary and tertiary structures.
The conformational changes in proteins were also confirmed in relatively low electric field strengths (<0.5 V/nm) and exposure for long periods of time. Bekard and Dunstan reported conformational change lysozyme in an AC low electric field of 10 Hz in a frequency with a range from 0.78 to 5.0 V/cm, as shown in Figure 184 [29][6]. The conformational changes are monitored with the time evolution of the relative emission intensity of lysozyme solutions at 346 nm of the tryptophan fluorescence emission with an excitation wavelength of 295 nm. The conformation during the first hour is monitored without being exposed to an AC electric field, shown by dotted vertical lines, followed by 3 h with AC electric field exposure and a further 2 h without the electric field again. In the experiment, it was confirmed that the tryptophan fluorescence emission not only decreased its intensity, but that the red shift of the emission wavelength peak was caused by exposure to the electric field. The spectral changes generally indicate alterations in the microenvironment of tryptophan residues, and typically reflect the exposure of these residues, initially concealed in hydrophobic segments of the folded protein, to the surrounding aqueous environment. The decrements of the relative fluorescence emission intensity of lysozyme are observed for exposure to all electric field strengths, and is more pronounced at a field strength of up to 5.0 V/cm. The decrease in tryptophan emission intensity appeared irreversible. Further analysis of the data indicates a linear relation between the relative tryptophan emission intensity and the applied electric field strength, as shown in Figure 195 [29][6].
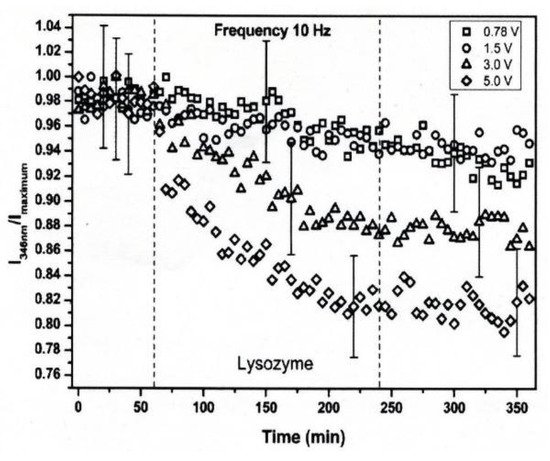
Figure 184. Time-evolution of the relative emission intensity of lysozyme solutions at 0.2 mg/mL (pH 7.2) monitored at 346 nm for exposure to varying electric field strengths. The electric field strengths are (□) 0.78, (○) 1.5, (△) 3.0 and (◇) 5.0 V/cm. The dotted lines indicate partitioning into the first 1 h without exposure to the electric field, followed by 3 h of electric field exposure and a further 2 h of without the electric field [29][6]. © Royal Society of Chemistry 2014. With permission of Royal Society of Chemistry.
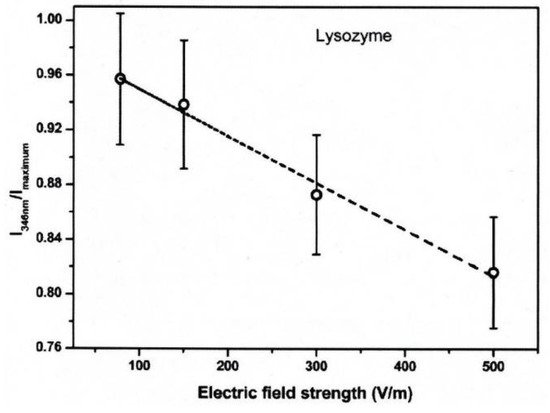
Figure 195. Relative fluorescence emission intensity of lysozyme solutions (0.2 mg/mL, pH 7.2) monitored at 346 nm as a function of electric field strength after 3 h of electric field exposure. The dotted line is a linear fit with an R2 of 0.99 [29][6]. © Royal Society of Chemistry 2014. With permission of Royal Society of Chemistry.
Bekard and Dunstan also reported that the fractions of the secondary structures of lysozyme solutions were changed from 31% α-helix, 20% β-strands, 20% β-turns and 29% random coils to 19% α-helix, 28% β-strands, 23% β-turns and 30% random coils after 3 h of exposure to an electric field of 3.0 V/cm strength. The electric field strength of 3.0 V/cm corresponds to be 0.1 fN of electrical force on HBs in the protein, which is almost six orders of magnitude lower than the HB bonding forces. To solve the inconsistency, Bekard and Dunstan proposed the model of indirect effect (slow process), which is based on the electrophoretic motion (electrostatic interactions) of a protein leading a frictional force for the protein unfolding [29][6]. The electrostatic effect is basically caused by oppositely charged terminal residues, charged side chains and peptide dipoles in the secondary structure segments of a protein. The dipole moment of lysozyme, which has a net charge of +7, is roughly calculated as 74 Debye length at natural pH. The alignment of secondary structure dipoles strongly affects the stability of the tertiary structure of proteins. In addition, the macro-dipole can distort the field distribution and produce relatively strong local electric fields. The electric field strength along a helix axis is estimated to be in the region of 109 V/m.
3. PEF Treatment for α-Amylase Inactivation via Conformational Changes
In some food processes, such as brewing and fermenting, the inactivation of enzymes is the final step before distributing food products to consumers. In the processing of frozen food of agricultural products, hot water treatments are commonly used as blanching, which is used for inactivating microorganisms and enzymes at the final stage of the process. PEF treatment is one of the candidates used to alternate non-thermal methods for enzyme inactivation instead of the thermal process. The PEF treatments for enzyme inactivation have been investigated by some researchers [36][13]. Yeom and Zhang confirmed that the functions of enzymatic proteins were inactivated by PEF treatment in some optimized circuit parameters [37][14]. Vega-Mercado et al. also reported that PEF parameters such as strength, pulse width, number of pulses and rise time of the pulse mainly affected the efficiency of enzyme inactivation. They also confirmed that inactivation of enzymes required generally more energy (i.e., PEF strength, pulse width, and number of pulses) than microorganisms did [38][15]. Castro et al. indicated that the pulse width of a PEF was more important for the inactivation of enzymes than PEF strength was. They confirmed that the enzymic protein of alkaline phosphatase in milk was inactivated by 65% in a PEF of 22 kV/cm strength, 0.7 msec width and 70 pulses, whereas it was not inactivated in a PEF of 26 kV/cm, 0.39 msec and 20 pulses [39][16]. Concerning the inactivation mechanism of enzymes by the PEF treatment, Dong et al. pointed out that the conformational changes in enzymic proteins such as denaturation and aggregation caused the inactivation of enzymic proteins [40][17].
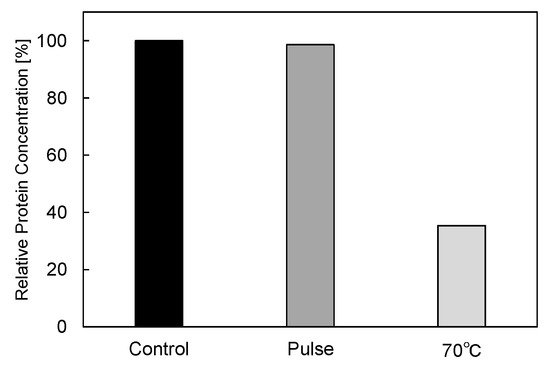
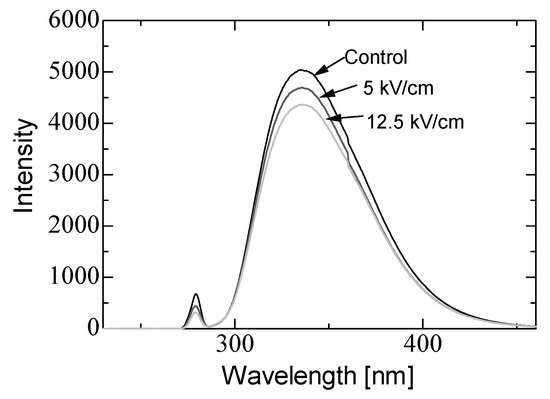
In basic experiments on enzyme inactivation by PEF treatment, small-scale vessels are sometimes used with parallel plane electrodes to generate homogeneous electric fields between the electrodes. Guionet et al. reported the effect of PEF treatment on enzymic inactivation of α-amylase. They developed a PFN circuit for controlling the pulse width and strength of a PEF, which was applied between the electrodes with a 4 mm gap in a cuvette, as shown in Figure 206 [6][18]. The cuvette was filled with α-amylase solution, which was prepared by dissolving 25 mg α-amylase in a solution consisting of 48 mL of distilled water and 2 mL of phosphate buffer. The results showed that the residual activity of α-amylase decreased with PEF strength at the same input energy with 10 μs of pulse width, as shown in Figure 217 [9][2]. This result indicates that the PEF strength strongly affects the efficiency of protein conformational change. They also confirmed conformational change in proteins due to PEF treatment, as shown in Figure 228 [9][2]. The tertiary structure change in α-amylase was monitored by fluorescence spectra at a 280 nm wavelength of excitation light. The tertiary structure (mainly tryptophan; Trp) of α-amylase also decreased with PEF strength at the same input energy. The enzymic active center of α-amylase was the carboxyl terminus of tryptophan. α-amylase commonly consists of three domains, which include several α-helix and β-sheet secondary structures. PEF treatments mainly affect hydrogen bonds in secondary structures (i.e., α-helix and β-sheet structures) and tertiary structures of α-amylase. It was also confirmed that the PEF and the heat treatments were different pathways for enzyme inactivation, as shown in Figure 239 [9][2]. They checked the aggregation of proteins after treatments by a PEF with 12.5 kV/cm and heating up to 70 °C. Both treatments caused the inactivation of α-amylase in same level of lower than 0.01 U/mL in residual activity. The relative protein concentration after filtering with a 0.22 μm syringe filter of PEF-treated α-amylase solution is almost same level as the control (without treatment), whereas the protein concentration of heat-treated α-amylase solution decreases to approximately 37%. The aggregation is confirmed to be only caused by heat treatment, because a decrease in the relative protein is caused by aggregation. Therefore, PEF treatment mainly contributes to conformational change in the protein, resulting in enzymatic activity change [6][18].
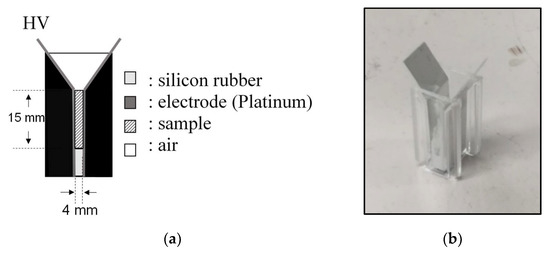
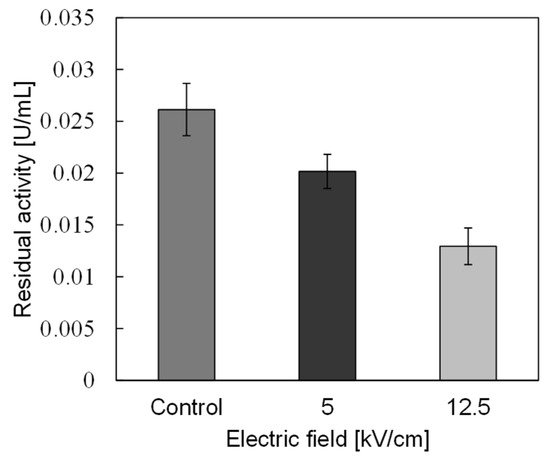
4. Enzyme Inactivation by a PEF under Various Conditions
Refolding denatured proteins has the potential to recover the enzymic activity of denatured enzymes, and this is an important issue for effective enzyme usage in the food industry [41][19]. PEF treatment induces the conformational change in proteins. Therefore, PEF treatment is a candidate for a novel method to refold the denatured enzymes. Ohshima et al. confirmed that the activity of six kinds of enzyme increased by a rate of 105%–120% via PEF treatment. They concluded that these enzyme activations were caused by protein conformational change or enzyme hydration [8][4]. They also reported that the activity of thermally denatured peroxidase recovered to 60% of its initial activity via PEF treatment with 12 kV/cm strength, 50 Hz repetition rate and 30 s exposure, whereas the enzymic activity recovered to only 40% of its initial activity at spontaneous refolding of the enzyme. However, the activity of thermally denatured lactate dehydrogenase (LDH) decreased due to PEF treatment, which suggested that further inactivation was caused by the application of a PEF to the thermally denatured LDH [8][4]. Therefore, the effect of PEF treatment on the refolding of the denatured proteins depends on the protein structure, i.e., the type of enzyme.
PEF treatment is a relatively low-temperature process compared to heat treatment, and is effective for the inactivation of not only foodborne and food spoilage bacteria but also enzymic proteins without degrading nutritional and sensory properties [42,43][20][21]. However, sometimes the PEF treatment alone for enzyme inactivation requires a long period of process time or significant electrical input power. Shamsi et al. proposed a combination of moderate heat treatment and PEF treatment to enhance the efficiency of inactivating enzymes and bacteria in whole milk [44][22]. Ho et al. confirmed that inactivating enzymes generally required more input energy in PEF treatment compared to the inactivation of microorganisms [45][23]. Agcam et al. conducted the inactivation of pectin methyl esterase (PME) in orange juice by PEF treatment. The inactivation of PME was significantly induced at a large input energy of PEF irradiation to the PME solution. A kinetic model was also proposed for estimating the efficiency of PME inactivation. In the model, the inactivation efficiency was expressed as a function of PEF treatment conditions, such as input power and treatment time. The kinetic model was confirmed to be effective for estimating the reaction rate and the time required for 90% inactivation [46][24]. Sharma et al. reported the effect of PEF treatment on the inactivation of four indigenous enzymes in whole milk. Lipase, plasmin, xanthine oxidase and alkaline phosphatase were used as specimens of indigenous enzymes in raw milk. The experimental results showed that the enzymic activities of plasmin, xanthine oxidase and lipolytic decreased with a 12%, 32%, and 82% reducing rate, respectively, compared to raw whole milk by a PEF treatment with 26.1 kV/cm strength at a 34 μs pulse width. When the strength of PEF increased to greater than 20.7 kV/cm for 34–101 μs, the enzymic activity of alkaline phosphatase was reduced to a comparable to thermal treatments. These results indicated that thermal effects also contribute to the inactivation of bacteria and enzymes along with the PEF treatments [47][25].
References
- Takaki, K.; Hayashi, N.; Wang, D.; Ohshima, T. High-voltage technologies for agriculture and food processing. J. Phys. D Appl. Phys. 2019, 52, 473001.
- Ohshima, T.; Tanino, T.; Guionet, A.; Takahashi, K.; Takaki, K. Mechanism of pulsed electric field enzyme activity change and pulsed discharge permeabilization of agricultural products. Jpn. J. Appl. Phys. 2021, 60, 060501.
- Syed, Q.A.; Ishaq, A.; Rahman, U.U.; Aslam, S.; Shukat, R. Pulsed electric field technology in food preservation: A review. J. Nutr. Health Food Eng. 2017, 6, 168–172.
- Ohshima, T.; Tamura, T.; Sato, M. Influence of pulsed electric field on various enzyme activities. J. Electrost. 2007, 65, 156–161.
- Mohamed, M.E.A.; Eissa, A.H.A. Pulsed electric fields for food processing technology. In Structure and Function of Food Engineering; Eissa, A.A., Ed.; Intech Open: London, UK, 2012; Chapter 11; pp. 275–304.
- Bekard, I.; Dunstan, D.E. Electric field induced changes in protein conformation. Soft Matter 2014, 10, 431–437.
- Jiang, Z.; You, L.; Dou, W.; Sun, T.; Xu, P. Effects of an Electric Field on the Conformational Transition of the Protein: A Molecular Dynamics Simulation Study. Polymers 2019, 11, 282.
- Qin, Z.; Buehler, M.J. Molecular Dynamics Simulation of the α-Helix to β-Sheet Transition in Coiled Protein Filaments: Evidence for a Critical Filament Length Scale. Phys. Rev. Lett. 2010, 104, 198304.
- Marracino, P.; Paffi, A.; Reale, R.; Liberti, M.; d’Inzeo, G.; Apollonio, F. Technology of High–Intensity Electric–Field Pulses: A Way to Control Protein Unfolding. J. Phys. Chem. Biophys. 2013, 3, 1000117.
- Valle, E.D.; Marracino, P.; Pakhomova, O.; Liberti, M.; Apollonio, F. Nanosecond pulsed electric signals can affect electrostatic environment of proteins below the threshold of conformational effects: The case study of SOD1 with a molecular simulation study. PLoS ONE 2019, 14, e0221685.
- Sheu, S.-Y.; Yang, D.-Y.; Selzle, H.L.; Schlag, E.W. Energetics of hydrogen bonds in peptides. Proc. Natl. Acad. Sci. USA 2003, 100, 12683–12687.
- Ding, F.; Borreguero, J.M.; Buldyrey, S.V.; Stanley, H.E.; Dokholyan, N.V. Mechanism for the α-Helix to β-Hairpin Transition. Proteins 2003, 53, 220–228.
- Van Loey, A.; Verachtert, B.; Hendrickx, M. Effects of high electric field pulses on enzymes. Trends Food Sci. Technol. 2002, 12, 94–102.
- Yeom, H.W.; Zhang, Q.H. Enzymatic inactivation by pulsed electric fields: A review. In Pulsed Electric Fields in Food Processing; Barbosa-Cánovas, G.V., Zhang, Q.H., Eds.; Technomic Publishing: Lancaster, UK, 2001; pp. 57–63.
- Vega-Mercado, H.M.; Martin-Belloso, O.; Qin, B.L.; Chang, F.J.; Góngora-Nieto, M.M.; Barbara-Cánovas, G.V.; Swanson, B.G. Non-thermal food preservation: Pulsed electric fields. Trends Food Sci. Technol. 1997, 8, 151–157.
- Castro, A.J.; Swanson, B.G.; Barbosa-Cánovas, G.V.; Zhang, Q.H. Pulsed electric fields modification of milk alkaline phosphatase activity. In Pulsed Electric Fields in Food Processing; Barbosa-Cánovas, G.V., Zhang, Q.H., Eds.; Technomic Publishing: Lancaster, UK, 2001; pp. 65–82.
- Dong, M.; Xu, Y.; Zhang, Y.; Han, M.; Wang, P.; Xu, X.; Zhou, G. Physicochemical and structural properties of myofibrillar proteins isolated from pale, soft, exudative (PSE)-like chicken breast meat: Effects of pulsed electric field (PEF). Innov. Food Sci. Emerg. Technol. 2020, 59, 102277.
- Guionet, A.; Fujiwara, T.; Sato, H.; Takahashi, K.; Takaki, K.; Matsui, M.; Tanino, T.; Ohshima, T. Pulsed electric fields act on tryptophan to inactivate α-amylase. J. Electrost. 2021, 112, 103597.
- Kubo, T.; Mizobata, T.; Kawata, Y. Refolding of yeast enolase in the presence of the chaperonin GroE. The nucleotide specificity of GroE and the role of GroES. J. Biol. Chem. 1993, 268, 19346–19351.
- Yeom, H.W.; Streaker, C.B.; Zhang, Q.H.; Min, D.B. Effects of pulsed electric fields on the activities of microorganisms and pectin methyl esterase in orange juice. J. Food Sci. 2000, 65, 1359–1363.
- Elez-Martínez, P.; Aguiló-Aguayo, I.; Martin-Belloso, O. Inactivation of orange juice peroxidase by high-intensity pulsed electric fields as influenced by process parameters. J. Sci. Food Agric. 2006, 86, 71–81.
- Shamsi, K.; Versteeg, C.; Sherkat, F.; Wan, J. Alkaline phosphatase and microbial inactivation by pulsed electric field in bovine milk. Innov. Food Sci. Emerg. Technol. 2008, 9, 217–223.
- Ho, S.Y.; Mittal, G.S.; Cross, J.D. Effects of high field electric pulses on the activity of selected enzymes. J. Food Eng. 1997, 31, 69–84.
- Agcam, E.; Akyıldız, A.; Evrendilek, G.A. Effects of PEF and heat pasteurization on PME activity in orange juice with regard to a new inactivation kinetic model. Food Chem. 2014, 165, 70–76.
- Sharma, P.; Oey, I.; Bremer, P.; Everett, D.W. Reduction of bacterial counts and inactivation of enzymes in bovine whole milk using pulsed electric fields. Int. Dairy J. 2014, 39, 146–156.
More