Protein glycosylation is a highly conserved post-translational modification among organisms. It plays fundamental roles in many biological processes, ranging from protein trafficking and cell adhesion to host–pathogen interactions. According to the amino acid side chain atoms to which glycans are linked, protein glycosylation can be divided into two major categories: N-glycosylation and O-glycosylation. However, there are other types of modifications such as the addition of GPI to the C-terminal end of the protein. Besides the importance of glycoproteins in biological functions, they are a major component of the fungal cell wall and plasma membrane and contribute to pathogenicity, virulence, and recognition by the host immunity. Given that this structure is absent in host mammalian cells, it stands as an attractive target for developing selective compounds for the treatment of fungal infections.
- N-glycosylation
- O-glycosylation
- glycosylphosphatidylinositol anchors
- host–fungus interaction
1. Introduction
2. The Candida albicans Protein Glycosylation Pathways
2.1. The N-Linked Glycosylation Pathway
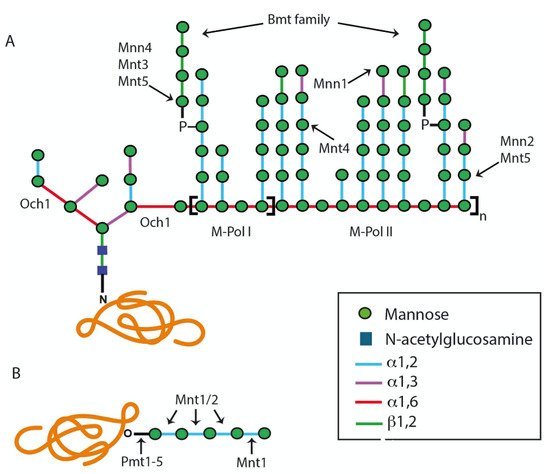
2.1.1. Assembly of Lipid-Linked Oligosaccharide
2.1.2. Modification of the N-Linked Glycan Core by Glycosidases and Transferases
2.2. The O-Linked Glycosylation Pathway
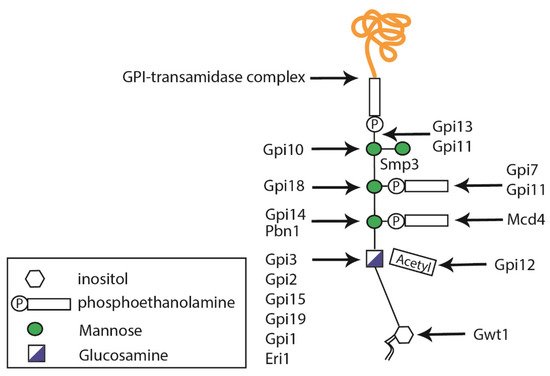
2.3. Glycosylphosphatidylinositol Anchor Synthesis
3. Protein Glycosylation in Other Medically Relevant Fungal Pathogens
References
- Lin, B.; Qing, X.; Liao, J.; Zhuo, K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells 2020, 9, 1022.
- Kornfeld, R.; Kornfeld, S. Assembly of Asparagine-Linked Oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664.
- Herscovics, A.; Orlean, P. Glycoprotein biosynthesis in yeast. FASEB J. 1993, 7, 540–550.
- Gavel, Y.; von Heijne, G.; Creaser, E.; Murali, C.; Britt, K. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. Des. Sel. 1990, 3, 433–442.
- Tanner, W.; Lehle, L. Protein glycosylation in yeast. Biochim. Biophys. Acta Rev. Biomembr. 1987, 906, 81–99.
- Yadav, U.; Khan, M.A. Targeting the GPI biosynthetic pathway. Pathog. Glob. Health. 2018, 112, 115–122.
- Lin, G.-Y.; Chang, C.-F.; Lan, C.-Y. The interaction between Carbohydrates and the Antimicrobial Peptide P-113Tri is Involved in the Killing of Candida albicans. Microorganisms 2020, 8, 299.
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 1–25.
- Díaz-Jiménez, D.F.; Pérez-García, L.A.; Martínez-Álvarez, J.A.; Mora-Montes, H.M. Role of the Fungal Cell Wall in Pathogenesis and Antifungal Resistance. Curr. Fungal Infect. Rep. 2012, 6, 275–282.
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78.
- Latgé, J.-P. Tasting the fungal cell wall. Cell. Microbiol. 2010, 12, 863–872.
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden Killers: Human Fungal Infections. Sci. Transl. Med. 2012, 4, 165rv13.
- Martinez-Duncker, I.; Díaz-Jímenez, D.F.; Mora-Montes, H.M. Comparative Analysis of Protein Glycosylation Pathways in Humans and the Fungal Pathogen Candida albicans. Int. J. Microbiol. 2014, 2014, 267497.
- Parodi, A.J. N-Glycosylation in trypanosomatid protozoa. Glycobiology 1993, 3, 193–199.
- Burda, P.; Aebi, M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta Gen. Subj. 1999, 1426, 239–257.
- Lehle, L.; Strahl, S.; Tanner, W. Protein Glycosylation, Conserved from Yeast to Man: A Model Organism Helps Elucidate Congenital Human Diseases. Angew. Chem. Int. Ed. 2006, 45, 6802–6818.
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta Bioenerg. 2013, 1833, 2430–2437.
- Dean, N. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta Gen. Subj. 1999, 1426, 309–322.
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582.
- Chojnacki, T.; Dallner, G. The biological role of dolichol. Biochem. J. 1988, 251, 1–9.
- Juchimiuk, M.; Kruszewska, J.; Palamarczyk, G. Dolichol phosphate mannose synthase from the pathogenic yeast Candida albicans is a multimeric enzyme. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 2265–2275.
- Samuelson, J.; Banerjee, S.; Magnelli, P.; Cui, J.; Kelleher, D.J.; Gilmore, R.; Robbins, P.W. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. USA 2005, 102, 1548–1553.
- Niewiadomska, M.; Janik, A.; Perlińska-Lenart, U.; Piłsyk, S.; Palamarczyk, G.; Kruszewska, J.S. The role of Alg13 N-acetylglucosaminyl transferase in the expression of pathogenic features of Candida albicans. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 789–801.
- Mora-Montes, H.M.; Ponce-Noyola, P.; Villagómez-Castro, J.C.; Gow, N.A.; Flores-Carreón, A.; López-Romero, E. Protein glycosylation in Candida. Futur. Microbiol. 2009, 4, 1167–1183.
- Yan, Q.; Lennarz, W.J. Oligosaccharyltransferase: A Complex Multisubunit Enzyme of the Endoplasmic Reticulum. Biochem. Biophys. Res. Commun. 1999, 266, 684–689.
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Díaz-Jiménez, D.F.; López-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.P.; Odds, F.C.; et al. Endoplasmic Reticulum α-Glycosidases of Candida albicans Are Required for N Glycosylation, Cell Wall Integrity, and Normal Host-Fungus Interaction. Eukaryot. Cell 2007, 6, 2184–2193.
- Nishikawa, A.; Poster, J.B.; Jigami, Y.; Dean, N. Molecular and Phenotypic Analysis of CaVRG4, Encoding an Essential Golgi Apparatus GDP-Mannose Transporter. J. Bacteriol. 2002, 184, 29–42.
- Bates, S.; Hughes, H.B.; Munro, C.; Thomas, W.P.; MacCallum, D.; Bertram, G.; Atrih, A.; Ferguson, M.; Brown, A.J.; Odds, F.C.; et al. Outer Chain N-Glycans Are Required for Cell Wall Integrity and Virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98.
- Mille, C.; Bobrowicz, P.; Trinel, P.-A.; Li, H.; Maes, E.; Guerardel, Y.; Fradin, C.; Martínez-Esparza, M.; Davidson, R.C.; Janbon, G.; et al. Identification of a New Family of Genes Involved in β-1,2-Mannosylation of Glycans in Pichia pastoris and Candida albicans. J. Biol. Chem. 2008, 283, 9724–9736.
- González-Hernández, R.J.; Jin, K.; Hernández-Chávez, M.J.; Díaz-Jiménez, D.F.; Trujillo-Esquivel, E.; Clavijo-Giraldo, D.M.; Tamez-Castrellón, A.K.; Franco, B.; Gow, N.A.R.; Mora-Montes, H.M. Phosphomannosylation and the Functional Analysis of the Extended Candida albicans MNN4-Like Gene Family. Front. Microbiol. 2017, 8, 2156.
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Díaz-Jiménez, D.F.; Kullberg, B.J.; Brown, A.J.; Odds, F.C.; et al. A Multifunctional Mannosyltransferase Family in Candida albicans Determines Cell Wall Mannan Structure and Host-Fungus Interactions. J. Biol. Chem. 2010, 285, 12087–12095.
- Díaz-Jiménez, D.F.; Mora-Montes, H.M.; Hernández-Cervantes, A.; Luna-Arias, J.P.; Gow, N.A.; Flores-Carreón, A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 2012, 419, 77–82.
- Goto, M. ProteinO-Glycosylation in Fungi: Diverse Structures and Multiple Functions. Biosci. Biotechnol. Biochem. 2007, 71, 1415–1427.
- Munro, C.; Bates, S.; Buurman, E.T.; Hughes, H.B.; MacCallum, D.; Bertram, G.; Atrih, A.; Ferguson, M.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans Are Partially Redundant α-1,2-Mannosyltransferases That Participate in O-Linked Mannosylation and Are Required for Adhesion and Virulence. J. Biol. Chem. 2005, 280, 1051–1060.
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 405–420.
- Newman, H.A.; Romeo, M.J.; Lewis, S.E.; Yan, B.C.; Orlean, P.; Levin, D.E. Gpi19, the Saccharomyces cerevisiae Homologue of Mammalian PIG-P, Is a Subunit of the Initial Enzyme for Glycosylphosphatidylinositol Anchor Biosynthesis. Eukaryot. Cell 2005, 4, 1801–1807.
- Grimme, S.J.; Colussi, P.A.; Taron, C.H.; Orlean, P. Deficiencies in the essential Smp3 mannosyltransferase block glycosylphosphatidylinositol assembly and lead to defects in growth and cell wall biogenesis in Candida albicans. Microbiology 2004, 150, 3115–3128.
- Peter, O.; Menon, A. Thematic review series: Lipid Posttranslational Modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011.
- Fraering, P.; Imhof, I.; Meyer, U.; Strub, J.-M.; Van Dorsselaer, A.; Vionnet, C.; Conzelmann, A. The GPI Transamidase Complex of Saccharomyces cerevisiae Contains Gaa1p, Gpi8p, and Gpi16p. Mol. Biol. Cell 2001, 12, 3295–3306.
- Kudoh, A.; Okawa, Y.; Shibata, N. Significant structural change in both O- and N-linked carbohydrate moieties of the antigenic galactomannan from Aspergillus fumigatus grown under different culture conditions. Glycobiology 2014, 25, 74–87.
- Latgé, J.P.; Mouyna, I.; Tekaia, F.; Beauvais, A.; Debeaupuis, J.P.; Nierman, W. Specific molecular features in the organization and biosynthesis of the cell wall ofAspergillus fumigatus. Med. Mycol. 2005, 43, 15–22.
- Tefsen, B.; Ram, A.F.J.; Van Die, I.; Routier, F. Galactofuranose in eukaryotes: Aspects of biosynthesis and functional impact. Glycobiology 2011, 22, 456–469.
- Katafuchi, Y.; Li, Q.; Tanaka, Y.; Shinozuka, S.; Kawamitsu, Y.; Izumi, M.; Ekino, K.; Mizuki, K.; Takegawa, K.; Shibata, N.; et al. GfsA is a β1,5-galactofuranosyltransferase involved in the biosynthesis of the galactofuran side chain of fungal-type galactomannan in Aspergillus fumigatus. Glycobiology 2017, 27, 568–581.
- Komachi, Y.; Hatakeyama, S.; Motomatsu, H.; Futagami, T.; Kizjakina, K.; Sobrado, P.; Ekino, K.; Takegawa, K.; Goto, M.; Nomura, Y.; et al. gfsAencodes a novel galactofuranosyltransferase involved in biosynthesis of galactofuranose antigen ofO-glycan inAspergillus nidulansandAspergillus fumigatus. Mol. Microbiol. 2013, 90, 1054–1073.
- Engel, J.; Schmalhorst, P.; Dörk, T.; Ferrieres, V.; Routier, F.H. A Single UDP-galactofuranose Transporter Is Required for Galactofuranosylation in Aspergillus fumigatus. J. Biol. Chem. 2009, 284, 33859–33868.
- Henry, C.; Li, J.; Danion, F.; Alcazar-Fuoli, L.; Mellado, E.; Beau, R.; Jouvion, G.; Latgé, J.-P.; Fontaine, T. Two KTR Mannosyltransferases Are Responsible for the Biosynthesis of Cell Wall Mannans and Control Polarized Growth in Aspergillus fumigatus. mBio 2019, 10, e02647-18.
- Onoue, T.; Tanaka, Y.; Hagiwara, D.; Ekino, K.; Watanabe, A.; Ohta, K.; Kamei, K.; Shibata, N.; Goto, M.; Oka, T. Identification of Two Mannosyltransferases Contributing to Biosynthesis of the Fungal-type Galactomannan α-Core-Mannan Structure in Aspergillus fumigatus. Sci. Rep. 2018, 8, 16918.
- Latgé, J.-P.; Beauvais, A.; Chamilos, G. The Cell Wall of the Human Fungal PathogenAspergillus fumigatus: Biosynthesis, Organization, Immune Response, and Virulence. Annu. Rev. Microbiol. 2017, 71, 99–116.
- Henry, C.; Fontaine, T.; Heddergott, C.; Robinet, P.; Aimanianda, V.; Beau, R.; Beauvais, A.; Mouyna, I.; Prevost, M.-C.; Fekkar, A.; et al. Biosynthesis of cell wall mannan in the conidium and the mycelium ofAspergillusfumigatus. Cell. Microbiol. 2016, 18, 1881–1891.
- Klutts, J.S.; Yoneda, A.; Reilly, M.C.; Bose, I.; Doering, T.L. Glycosyltransferases and their products: Cryptococcal variations on fungal themes. FEMS Yeast Res. 2006, 6, 499–512.
- Rodrigues, M.; Dobroff, A.S.S.; Couceiro, J.N.D.S.S.; Alviano, C.S.; Schauer, R.; Travassos, L.R. Sialylglycoconjugates and sialyltransferase activity in the fungus Cryptococcus neoformans. Glycoconj. J. 2002, 19, 165–173.
- Doering, T.L. How Sweet it is! Cell Wall Biogenesis and Polysaccharide Capsule Formation inCryptococcus neoformans. Annu. Rev. Microbiol. 2009, 63, 223–247.
- Schutzbach, J.; Ankel, H.; Brockhausen, I. Synthesis of cell envelope glycoproteins of Cryptococcus laurentii. Carbohydr. Res. 2007, 342, 881–893.
- Lee, D.-J.; Bahn, Y.-S.; Kim, H.-J.; Chung, S.-Y.; Kang, H.A. Unraveling the Novel Structure and Biosynthetic Pathway of O-Linked Glycans in the Golgi Apparatus of the Human Pathogenic Yeast Cryptococcus neoformans. J. Biol. Chem. 2015, 290, 1861–1873.
- Reilly, M.; Aoki, K.; Wang, Z.A.; Skowyra, M.; Williams, M.; Tiemeyer, M.; Doering, T.L. A Xylosylphosphotransferase of Cryptococcus neoformans Acts in Protein O-Glycan Synthesis. J. Biol. Chem. 2011, 286, 26888–26899.
- Bezerra, L.L.L.-B.L. Sporothrix schenckii Cell Wall Peptidorhamnomannans. Front. Microbiol. 2011, 2, 243.
- Tamez-Castrellón, A.K.; van der Beek, S.L.; López-Ramírez, L.A.; Martínez-Duncker, I.; Lozoya-Pérez, N.E.; van Sorge, N.M.; Mora-Montes, H.M. Disruption of protein rhamnosylation affects the Sporothrix schenckii-host interaction. Cell Surf. 2021, 7, 100058.
- Lopes-Bezerra, L.M.; Mora-Montes, H.M.; Zhang, Y.; Nino-Vega, G.; Rodrigues, A.M.; De Camargo, Z.P.; De Hoog, S. Sporotrichosis between 1898 and 2017: The evolution of knowledge on a changeable disease and on emerging etiological agents. Med. Mycol. 2018, 56, S126–S143.
