Sulfate-reducing bacteria (SRB) are a heterogeneous group of anaerobic microorganisms that play an important role in producing hydrogen sulfide not only in the natural environment, but also in the gastrointestinal tract and oral cavity of animals and humans.
- anaerobic microorganisms
- sulfate-reducing bacteria
- hydrogen sulfide
- toxicity
1. Introduction
Sulfate-reducing bacteria (SRB) are microorganisms that occur in different ecosystems globally [1,2,3,4,5][1][2][3][4][5]. They can also be isolated from the gastrointestinal tract and the oral cavity of humans and animals [6,7,8,9,10,11][6][7][8][9][10][11]. The cultivation of SRB is sometimes fastidious, as they require anaerobic conditions, strict temperature regulations and precise pH requirements [12]. Consequently, research on SRB is uncommon, and a method of long-term cryopreservation has not been thoroughly developed. Therefore, in this work we review methods for cryopreservation and their application for preservation of SRB [13,14][13][14].
For cryopreservation, it is necessary to choose the right laboratory equipment in which long-term storage can be performed [15]. Another important step in cryopreservation is to select the right cryoprotectant to maximize the viability of the microorganism after freezing [13]. Although the type and concentration of the chosen cryoprotectant is critical, the possibility to combine different preservation compounds to achieve successful cryopreservation may be equally important. The viability of microorganisms is influenced by a number of factors [16]. Colony age, amount of inoculum, cell size or rate of cooling may impact the survival of the culture. Furthermore, viability can also vary between individual species within the same genus [13].
2. Sulfate-Reducing Bacteria in Various Biotopes
-
Gram-negative mesophilic SRB; these do not form spores and are one of the most widespread SRB in nature (genera Desulfovibrio, Desulfobotulus, Desulfobulbus, Desulfohalobium and Desulfomicrobium);
-
Gram-positive spore-forming bacteria; these are a typical representative (genus Desulfotomaculum) that can be identified from soil samples (according to the updated classification, these microorganisms are represented and included in order Clostridiales);
-
Gram-negative thermophilic sulfate-reducing microorganisms (genus Thermodesulfobacterium);
-
Gram-negative thermophilic archaeal sulfate-reducing microorganisms; these include members of the genus Archaeoglobus that can only be found in anaerobic, underwater, hydrothermal environments because they require salt and high temperature for their growth.
2.1. Water Environment
2.2. Surfaces of Corrosive Metals
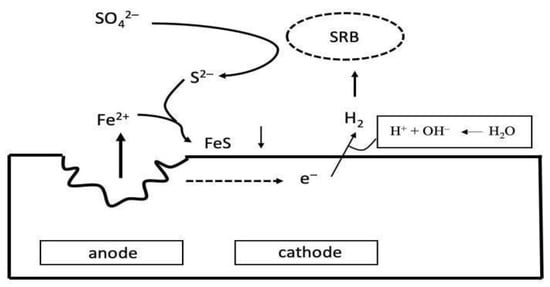
2.3. Corrosion of Concrete, Stone Elements and Masonry
2.4. Gastrointestinal Tract
3. Conditions Determining the Viability of Sulfate-Reducing Bacteria
3.1. Physical Conditions
3.2. Competition and Coexistence with Other Intestinal Microorganisms
3.3. Biochemical Characteristics of Sulfate-Reducing Bacteria
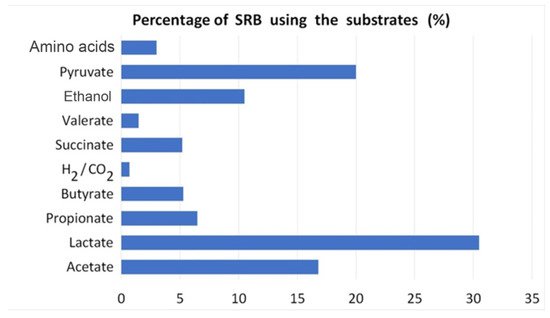
References
- Cambridge University Press. Sulphate-Reducing Bacteria: Environmental and Engineered Systems; Barton, L., Hamilton, W.A., Eds.; Cambridge University Press: Cambridge, NY, USA, 2007; ISBN 978-0-521-85485-6.
- Barton, L.L.; Fauque, G.D. Chapter 2 Biochemistry, Physiology and Biotechnology of Sulfate-Reducing Bacteria. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 68, pp. 41–98. ISBN 978-0-12-374803-4.
- Kushkevych, I. Isolation and Purification of Sulfate-Reducing Bacteria. In Microorganisms; Blumenberg, M., Shaaban, M., Elgaml, A., Eds.; IntechOpen: London, UK, 2020; ISBN 978-1-83880-187-8.
- Kushkevych, I.; Dordević, D.; Kollár, P. Analysis of Physiological Parameters of Desulfovibrio Strains from Individuals with Colitis. Open Life Sci. 2019, 13, 481–488.
- Kushkevych, I.; Dordević, D.; Vítězová, M. Analysis of PH Dose-Dependent Growth of Sulfate-Reducing Bacteria. Open Med. 2019, 14, 66–74.
- Kushkevych, I.; Kollar, P.; Suchy, P.; Parak, T.; Pauk, K.; Imramovsky, A. Activity of Selected Salicylamides against Intestinal Sulfate-Reducing Bacteria. Neuro Endocrinol. Lett. 2015, 36 (Suppl. 1), 106–113.
- Kushkevych, I.V. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio Piger Vib-7 and Desulfomicrobium Sp. Rod-9. Pol. J. Microbiol. 2015, 64, 107–114.
- Kushkevych, I.; Fafula, R.; Parák, T.; Bartoš, M. Activity of Na+/K+-Activated Mg2+-Dependent ATP-Hydrolase in the Cell-Free Extracts of the Sulfate-Reducing Bacteria Desulfovibrio Piger Vib-7 and Desulfomicrobium Sp. Rod-9. Acta Vet. Brno 2015, 84, 3–12.
- Kushkevych, I.V. Activity and Kinetic Properties of Phosphotransacetylase from Intestinal Sulfate-Reducing Bacteria. Acta Biochim. Pol. 2015, 62, 103–108.
- Kushkevych, I.; Coufalová, M.; Vítězová, M.; Rittmann, S.K.-M.R. Sulfate-Reducing Bacteria of the Oral Cavity and Their Relation with Periodontitis—Recent Advances. JCM 2020, 9, 2347.
- Ran, S.; Mu, C.; Zhu, W. Diversity and Community Pattern of Sulfate-Reducing Bacteria in Piglet Gut. J. Animal Sci. Biotechnol. 2019, 10, 40.
- Postgate, J. The Suphate-Reducing Bacteria, 2nd ed.; Cambridge University: Cambridge, NY, USA, 1984.
- Hubálek, Z. Cryopreservation of Microorganisms at Ultra-Low Temperatures; Elsevier: Amsterdam, The Netherlands, 1996.
- Trsic-Milanovic, N.; Kodzic, A.; Baras, J.; Dimitrijevic-Brankovic, S. The Influence of a Cryoprotective Medium Containing Glycerol on the Lyophilization of Lactic Acid Bacteria. J. Serb. Chem. Soc. 2001, 66, 435–441.
- Butterfield, W.; Jong, S.C.; Alexander, M.T. Polypropylene Vials for Preserving Fungi in Liquid Nitrogen. Mycologia 1978, 70, 1122–1124.
- Grout, B.; Morris, J.; Mclellan, M. Cryopreservation and the Maintenance of Cell Lines. Trends Biotechnol. 1990, 8, 293–297.
- Castro, H.F.; Williams, N.H.; Ogram, A. Phylogeny of Sulfate-Reducing Bacteria1. FEMS Microbiol. Ecol. 2000, 31, 1–9.
- Brenner, D.J.; Krieg, N.R.; Staley, J.T.; Garrity, G.M. The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. In Bergey’s Manual of Systematic Bacteriology; Springer: Boston, MA, USA, 2005; p. 1388.
- Kushkevych, I.; Vítězová, M.; Kos, J.; Kollár, P.; Jampílek, J. Effect of Selected 8-Hydroxyquinoline-2-Carboxanilides on Viability and Sulfate Metabolism of Desulfovibrio Piger. J. Appl. Biomed. 2018, 16, 241–246.
- Kushkevych, I.; Cejnar, J.; Treml, J.; Dordević, D.; Kollar, P.; Vítězová, M. Recent Advances in Metabolic Pathways of Sulfate Reduction in Intestinal Bacteria. Cells 2020, 9, 698.
- Kushkevych, I.; Kováč, J.; Vítězová, M.; Vítěz, T.; Bartoš, M. The Diversity of Sulfate-Reducing Bacteria in the Seven Bioreactors. Arch. Microbiol. 2018, 200, 945–950.
- Abdulina, D.; Kováč, J.; Iutynska, G.; Kushkevych, I. ATP Sulfurylase Activity of Sulfate-Reducing Bacteria from Various Ecotopes. Biotech 2020, 10, 55.
- Fauque, G.D. Ecology of Sulfate-Reducing Bacteria. In Sulfate-Reducing Bacteria; Barton, L.L., Ed.; Springer: Boston, MA, USA, 1995; pp. 217–241. ISBN 978-1-4899-1584-9.
- Hao, O.J.; Chen, J.M.; Huang, L.; Buglass, R.L. Sulfate-reducing Bacteria. Crit. Rev. Environ. Sci. Technol. 1996, 26, 155–187.
- Dordević, D.; Jančíková, S.; Vítězová, M.; Kushkevych, I. Hydrogen Sulfide Toxicity in the Gut Environment: Meta-Analysis of Sulfate-Reducing and Lactic Acid Bacteria in Inflammatory Processes. J. Adv. Res. 2020, 27, 55–69.
- Kushkevych, I.; Dordević, D.; Vítězová, M. Possible Synergy Effect of Hydrogen Sulfide and Acetate Produced by Sulfate-Reducing Bacteria on Inflammatory Bowel Disease Development. J. Adv. Res. 2020, 21, 71–78.
- Černý, M.; Vítězová, M.; Vítěz, T.; Bartoš, M.; Kushkevych, I. Variation in the Distribution of Hydrogen Producers from the Clostridiales Order in Biogas Reactors Depending on Different Input Substrates. Energies 2018, 11, 3270.
- Kováč, J.; Vítězová, M.; Kushkevych, I. Metabolic Activity of Sulfate-Reducing Bacteria from Rodents with Colitis. Open Med. 2018, 13, 344–349.
- Kushkevych, I.; Vítězová, M.; Fedrová, P.; Vochyanová, Z.; Paráková, L.; Hošek, J. Kinetic Properties of Growth of Intestinal Sulphate-Reducing Bacteria Isolated from Healthy Mice and Mice with Ulcerative Colitis. Acta Vet. Brno 2017, 86, 405–411.
- Plugge, C.M.; Zhang, W.; Scholten, J.C.M.; Stams, A.J.M. Metabolic Flexibility of Sulfate-Reducing Bacteria. Front. Microbio. 2011, 2, 81.
- Köpke, B.; Wilms, R.; Engelen, B.; Cypionka, H.; Sass, H. Microbial Diversity in Coastal Subsurface Sediments: A Cultivation Approach Using Various Electron Acceptors and Substrate Gradients. AEM 2005, 71, 7819–7830.
- Okabe, S.; Itoh, T.; Satoh, H.; Watanabe, Y. Analyses of Spatial Distributions of Sulfate-Reducing Bacteria and Their Activity in Aerobic Wastewater Biofilms. Appl. Environ. Microbiol. 1999, 65, 5107–5116.
- Newport, P.J.; Nedwell, D.B. The Mechanisms of Inhibition of Desulfovibrio and Desulfotomaculum Species by Selenate and Molybdate. J. Appl. Bacteriol. 1988, 65, 419–423.
- Islander, R.L.; Devinny, J.S.; Mansfeld, F.; Postyn, A.; Shih, H. Microbial Ecology of Crown Corrosion in Sewers. J. Environ. Eng. 1991, 117, 751–770.
- Leclerc, H.; Oger, C.; Beerens, H.; Mossel, D.A.A. Occurrence of Sulphate Reducing Bacteria in the Human Intestinal Flora and in the Aquatic Environment. Water Res. 1980, 14, 253–256.
- Gibson, G.R.; Macfarlane, G.T.; Cummings, J.H. Occurrence of Sulphate-Reducing Bacteria in Human Faeces and the Relationship of Dissimilatory Sulphate Reduction to Methanogenesis in the Large Gut. J. Appl. Bacteriol. 1988, 65, 103–111.
- Gibson, G.R.; Macfarlane, S.; Macfarlane, G.T. Metabolic Interactions Involving Sulphate-Reducing and Methanogenic Bacteria in the Human Large Intestine. FEMS Microbiol. Ecol. 1993, 12, 117–125.
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Bartoš, M. Production of Biogas: Relationship between Methanogenic and Sulfate-Reducing Microorganisms. Open Life Sci. 2017, 12, 82–91.
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Kováč, J.; Kaucká, P.; Jesionek, W.; Bartoš, M.; Barton, L. A New Combination of Substrates: Biogas Production and Diversity of the Methanogenic Microorganisms. Open Life Sci. 2018, 13, 119–128.
- Gajdács, M.; Spengler, G.; Urbán, E. Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik’s Cube of Clinical Microbiology? Antibiotics 2017, 6, 25.
- Gajdács, M.; Urbán, E. Relevance of Anaerobic Bacteremia in Adult Patients: A Never-Ending Story? Eur. J. Microbiol. Immunol. 2020, 10, 64–75.
- Gajdács, M.; Ábrók, M.; Lázár, A.; Terhes, G.; Urbán, E. Anaerobic Blood Culture Positivity at a University Hospital in Hungary: A 5-Year Comparative Retrospective Study. Anaerobe 2020, 63, 102200.
- Gibson, G.R.; Macfarlane, G.T. Chemostat Enrichment of Sulphate-Reducing Bacteria from the Large Gut. Lett. Appl. Microbiol. 1988, 7, 127–133.
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Growth and Activities of Sulphate-Reducing Bacteria in Gut Contents of Healthy Subjects and Patients with Ulcerative Colitis. FEMS Microbiol. Lett. 1991, 86, 103–112.
- Kushkevych, I.; Dordević, D.; Kollar, P.; Vítězová, M.; Drago, L. Hydrogen Sulfide as a Toxic Product in the Small-Large Intestine Axis and Its Role in IBD Development. JCM 2019, 8, 1054.
- Kushkevych, I.; Kotrsová, V.; Dordević, D.; Buňková, L.; Vítězová, M.; Amedei, A. Hydrogen Sulfide Effects on the Survival of Lactobacilli with Emphasis on the Development of Inflammatory Bowel Diseases. Biomolecules 2019, 9, 752.
- Kushkevych, I.; Castro Sangrador, J.; Dordević, D.; Rozehnalová, M.; Černý, M.; Fafula, R.; Vítězová, M.; Rittmann, S.K.-M.R. Evaluation of Physiological Parameters of Intestinal Sulfate-Reducing Bacteria Isolated from Patients Suffering from IBD and Healthy People. JCM 2020, 9, 1920.
- Florin, T.H.J.; Neale, G.; Goretski, S.; Cummings, J.H. The Sulfate Content of Foods and Beverages. J. Food Compos. Anal. 1993, 6, 140–151.
- Kováč, J.; Kushkevych, I. New Modification of Cultivation Medium for Isolation and Growth of Intestinal Sulfate-Reducing Bacteria. In Proceedings of the 24th International Ph.D. Students Conference, Brno, Czech Republic, 8–9 November 2017; Volume 2017, pp. 702–707.
- Fallingborg, J.; Christensen, L.A.; Ingeman-Nielsen, M.; Jacobsen, B.A.; Abildgaard, K.; Rasmussen, H.H. PH-Profile and Regional Transit Times of the Normal Gut Measured by a Radiotelemetry Device. Aliment. Pharmacol. Ther. 2007, 3, 605–614.
- Beerens, H.; Romond, C. Sulfate-Reducing Anaerobic Bacteria in Human Feces. Am. J. Clin. Nutr. 1977, 30, 1770–1776.
- Fite, A. Identification and Quantitation of Mucosal and Faecal Desulfovibrios Using Real Time Polymerase Chain Reaction. Gut 2004, 53, 523–529.
- Hilton, B.L.; Oleszkiewicz, J.A. Sulfide-Induced Inhibition of Anaerobic Digestion. J. Environ. Eng. 1988, 114, 1377–1391.
- Christl, S.U.; Gibson, G.R.; Cummings, J.H. Role of Dietary Sulphate in the Regulation of Methanogenesis in the Human Large Intestine. Gut 1992, 33, 1234–1238.
- Deplancke, B.; Hristova, K.R.; Oakley, H.A.; McCracken, V.J.; Aminov, R.; Mackie, R.I.; Gaskins, H.R. Molecular Ecological Analysis of the Succession and Diversity of Sulfate-Reducing Bacteria in the Mouse Gastrointestinal Tract. Appl. Environ. Microbiol. 2000, 66, 2166–2174.
- Kushkevych, I.; Dordević, D.; Vítězová, M.; Kollár, P. Cross-Correlation Analysis of the Desulfovibrio Growth Parameters of Intestinal Species Isolated from People with Colitis. Biologia 2018, 73, 1137–1143.
- Postgate, J.R. On the Nutrition of Desulphovibrio Desulphuricans. J. Gen. Microbiol. 1951, 5, 714–724.
- Postgate, J.R. Sulphate Reduction by Bacteria. Annu. Rev. Microbiol. 1959, 13, 505–520.
- Postgate, J.R.; Campbell, L.L. Classification of Desulfovibrio Species, the Nonsporulating Sulfate-Reducing Bacteria. Bacteriol. Rev. 1966, 30, 732–738.
- Bade, K.; Manz, W.; Szewzyk, U. Behavior of Sulfate Reducing Bacteria under Oligotrophic Conditions and Oxygen Stress in Particle-Free Systems Related to Drinking Water. FEMS Microbiol. Ecol. 2000, 32, 215–223.
- Krekeler, D.; Sigalevich, P.; Teske, A.; Cypionka, H.; Cohen, Y. A Sulfate-Reducing Bacterium from the Oxic Layer of a Microbial Mat from Solar Lake (Sinai), Desulfovibrio oxyclinae Sp. Nov. Arch. Microbiol. 1997, 167, 369–375.
- Attene-Ramos, M.S.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. Evidence That Hydrogen Sulfide Is a Genotoxic Agent. Mol. Cancer Res. 2006, 4, 9–14.
- Kushkevych, I.; Leščanová, O.; Dordević, D.; Jančíková, S.; Hošek, J.; Vítězová, M.; Buňková, L.; Drago, L. The Sulfate-Reducing Microbial Communities and Meta-Analysis of Their Occurrence during Diseases of Small–Large Intestine Axis. JCM 2019, 8, 1656.
- Kushkevych, I.; Dordević, D.; Vítězová, M. Toxicity of Hydrogen Sulfide toward Sulfate-Reducing Bacteria Desulfovibrio Piger Vib-7. Arch. Microbiol. 2019, 201, 389–397.
- Pitcher, M.C.; Cummings, J.H. Hydrogen Sulphide: A Bacterial Toxin in Ulcerative Colitis? Gut 1996, 39, 1–4.
