LINE-1 (L1) is a class of autonomous mobile genetic elements that form somatic mosaicisms in various tissues of the organism. The activity of L1 retrotransposons is strictly controlled by many factors in somatic and germ cells at all stages of ontogenesis. Alteration of L1 activity was noted in a number of diseases: in neuropsychiatric and autoimmune diseases, as well as in various forms of cancer.
- LINE-1 retrotransposons
- L1 silencing
- repetitive elements
- regulation
- Introduction
1. Introduction
Dispersed DNA repeats of LINE-1 (L1) retrotransposons account for 17% of the human genome [1]. Most of L1, which includes more than 500 thousand copies, is not active in the genome as they are truncated repeats or contain mutations in the protein-coding sequence necessary for retrotransposition [2]. However, approximately 150 copies are full-length and capable of self-copying and distributing in the genome [3,4,5]. Moreover, L1 elements provoke the spread of other genetic repeats, such as Alu and SVA [6,7,8]. Insertions of L1 elements occur mainly in the non-coding regions of the genome: introns and intergenic spaces [9]. The presence of the L1 element at a certain locus can affect gene expression and even lead to the formation of alternative transcripts, which can make a significant contribution to the functions of individual cells, tissues, and the whole organism [10,11].
- L1 Structure
2. L1 Structure
Figure 1) [12,13,14]. ORF1 encodes a ~40 kD protein with the chaperone activity necessary to stabilize a new L1 copy [15,16,17,18]. ORF2 encodes ~150 kD proteins with the endonuclease and retrotransposase activity required for the retrotransposition process [15,19,20]. In the opposite direction, ORF0 and two antisense promoters are located in the 5′and 3′ UTR. The function of ORF0 remains poorly understood. According to some data, ORF0 is involved in the formation of chimeric proteins or enhances the mobility of L1 [13,21,22,23].
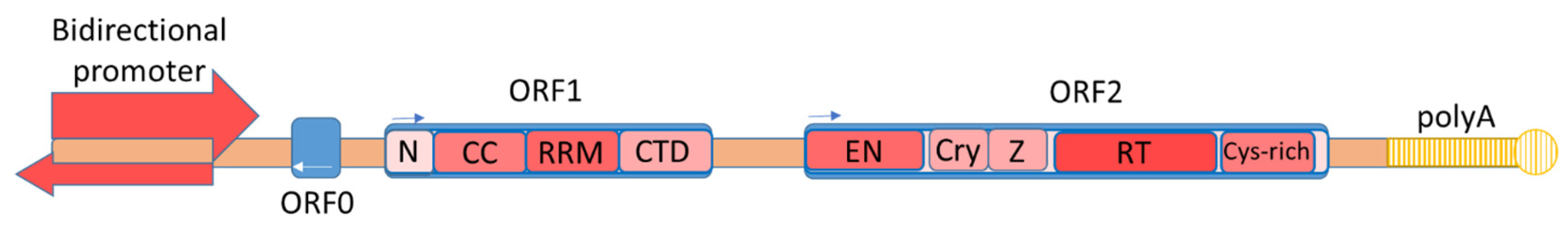
Figure 1. The structure of a full-length copy of L1 retrotransposon. ORF1 consists of an N-terminal domain (N), a coiled-coil domain (CCD), an RNA recognition motive (RRM), and a C-terminal domain (CTD) [18]. ORF2 consists of endonuclease (EN), retrotransposase (RT), a cryptic domain (Cry), a Z-domain (Z), and a C-terminal domain with a cysteine-rich region (Cys-rich) [24].
- Retrotransposition Mechanism
3. Retrotransposition Mechanism
Figure 2) [20,25,26]. A new RNA copy of the L1 element is expressed from the forward chain of the L1 element due to the strong promoter in the 5′UTR [14,27]. The new copy is then polyadenylated and leaves the nucleus [28]. Translation of ORF1 and ORF2 as well as the formation of L1 ribonucleoprotein (L1 RNP) occur in the cytoplasm. L1 RNP is a bicistronic mRNA coated with ORF1 proteins and contains one or two copies of the ORF2 protein [29]. In the cytoplasm, polyadenylate-binding protein 1 (PABPC1) attaches to the polyA tail of L1; its presence is critical for the formation of L1 RNP [28]. Transportation of L1 RNP from the cytoplasm to the nucleus is accomplished using the membrane-associated endosomal sorting complex required for transportation (ESCRT) [30]. It has also recently been shown that the ORF1 protein interacts with the KPNA2 and KPNB1 karyopherins, as well as possibly other KPNA family proteins involved in nuclear protein importation through nuclear pores (nuclear pore complex) [18]. The cancer cell model showed that L1 RNP penetrates the nucleus during mitosis and the integration of the new copy of the L1 element into the genome occurs in the S phase of a cell cycle [31]. Interestingly, in different tissues, the association of the L1 retrotransposition process with certain stages of the cell cycle can be different. For example, in neuronal cell cultures, it has been shown that retrotransposition can occur in non-dividing cells [32]. In entering the nucleus and reaching the genomic DNA, the endonuclease recognizes the consensus cleavage site 5′-TTTT/AA-3′ [20,33,34,35], and creates a single-stranded DNA break with the formation of both phosphate 5′-PO4 and hydroxyl 3′-OH groups at the ends [20]. The L1 transcript is attached via the polyA tail to the region of the endonuclease recognition site and reverse transcription of the L1 RNA occurs [36,37,38]. For the classical mechanism of retrotransposition, host proteins involved in DNA repair and replication are necessary [34,39,40]. A complex of PARP1 and PARP2 proteins is formed at the single-strand DNA breaks [40]. PARP2 specifically recognizes a single-stranded DNA gap at the L1 integration site. PARP2 is activated by the poly-ADP ribosylation process (PARylation). Activated PARP2 interacts with the RPA complex, which allows for the integration of a new synthesized L1 copy into the DNA. RPA, a replicative complex of protein A (heterotrimeric protein A complex), consisting of RPA70, RPA32, and RPA14 proteins, is required to bind single-stranded DNA in eukaryotes and to protect it from the deamination of cytidine [41]. The role of PARP1 in L1 retrotransposition has not been fully understood but it has been revealed that PARP1 interacts directly with ORF2, and the retrotransposase domain is responsible for this process. The absence of one of the PARP1 or PARP2 proteins leads to a decrease in the retrotransposition by about 50%; the absence of both proteins or the RPA complex reduces L1 retrotransposition by 80% [40]. The ORF2 complex, which is formed in the region of integration into the genome and promotes reverse transcription, includes various proteins involved in DNA stabilization and enzyme processability [42]. The first of these proteins is the proliferating cell nuclear antigen factor (PCNA). ORF2 interacts with PCNA and this interaction is critical for retrotransposition [43]. RUVBL1 and RUVBL2 repair proteins are also required for L1 spreading and their absence leads to a decrease in the retrotransposition [43]. In addition, the nonsense-mediated decay factor UPF1 and the MOV10 helicase were detected in the L1 RNP. Interestingly, UPF1 knockdown increases the amount of mRNA and L1 proteins but simultaneously reduces the effectiveness of the retrotransposition [43]. Inhibitory activity against L1 was shown for MOV10. However, in a recent study, it was suggested that MOV10 may facilitate the attachment of UPF1 to L1 RNP [42,44]. Insertion of the full-length L1 copy is a rare event; usually, a new copy of the L1 element is truncated from the 5′UTR [26,45,46,47]. Double-stranded DNA repair factors XRCC6 (Ku70/Ku80), Artemis (DCLRE1C), and LigIV (LIG4) are involved in the truncation of a new copy of the L1 element [46] (
Figure 2). The exact mechanism of their action remains unclear. It is suggested that XRCC6 can facilitate the attachment of ORF2 to the overlying targeted DNA, thus accelerating the completion of the retrotransposition process and leading to truncation [46]. After integration of the reverse complement strand of the new L1 copy, the second strand of DNA breaks and the first strand of the new L1 copy is synthesized using the host cellular enzymes involved in both DNA replication and reparation [34,40]. Retrotransposition can occur using an alternative endonuclease-independent (EN-independent) mechanism in p53-defective cells or cells containing mutations in the non-homologous end-junction (NHEJ) genes during DNA repair, which apparently uses DNA breaks to initiate transcription [45,48,49,50].
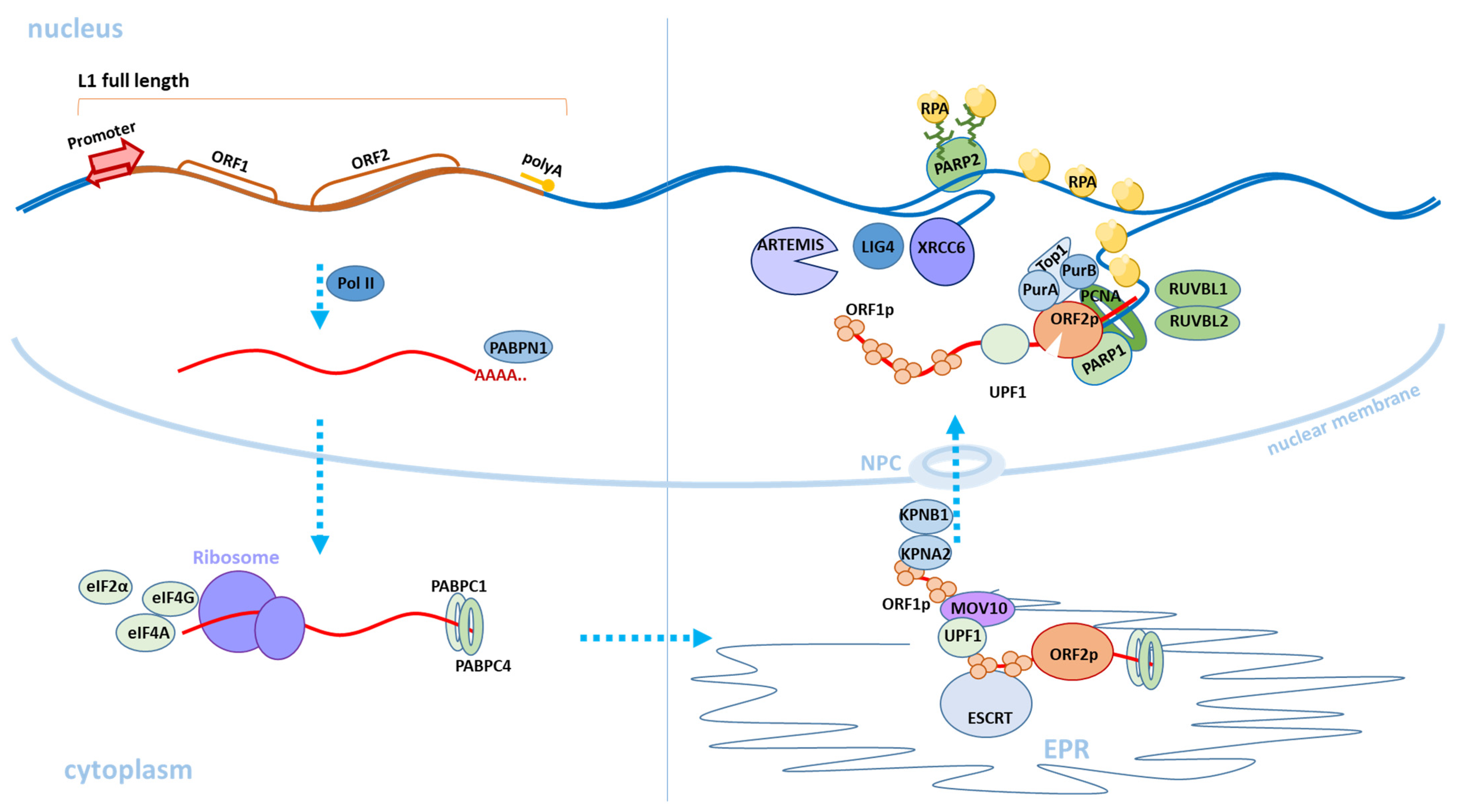
Figure 2. Scheme of the classical retrotransposition mechanism of L1. The transition of L1 from one stage of retrotransposition to another is indicated by blue dashed arrows. The upper left part of the figure shows the expression of a full-length copy of active L1 in the cell nucleus. The L1 RNA transcript (marked in red) is transported to the cytoplasm. The L1 ORF1p and ORF2p proteins are synthesized and the L1 RNP is formed (in the lower right part of the figure). Then, through the endoplasmic reticulum (EPR) and nuclear pore complex (NPC), L1 RNP is transported to the nucleus and L1 DNA copy formed by a reverse transcription is integrated into a new genomic locus (in the upper right part of the figure). The cellular factors involved in the retrotransposition process, which are described in this review, are also depicted.
- L1 Evolution
4. L1 Evolution
L1s belonging to the LINE class of mobile genetic elements are found in the genomes of animals and plants [51,52]. Although animal L1 is found in the genomes of some protostomes, the history of gradual accumulation and the evolution of modern L1s can be traced at the level of deuterostomes, possessed by three highly divergent groups: a united species from echinoderms to teleost fishes; non-mammal vertebrates; vertebrates from fish to mammals [52,53].
4.1. LINE Evolution in Deuterostomes and Non-Mammals
4.1. LINE Evolution in Deuterostomes and Non-Mammals
A high diversity of ancient L1 families was found in the lancelet, sea urchin, and tunicates, and despite their variability, they make up a small proportion of the repeated sequences [53,54]. Mobile elements are in constant competition with each other and with factors limiting their activity in the cell, and they try to maintain the ability to spread in the genome and increase the number of copies by capturing new genomic loci. However, this battle is not always successful considering that in most bony fish genomes, although there are exceptions, the number of LINEs is not large compared to other mobile elements, yielding to DNA transposons [54,55,56]. On the contrary, in the known representatives of living jawless fishes, cartilaginous fish, coelacanths, and lungfishes, the number of LINEs is not inferior to other classes of DNA repeats and constitutes 25–50% of all repeats in the genome [54,57,58,59,60,61,62]. Interestingly, increased diversity of the L1 families is observed in fish [53]. Moreover, the highest diversity of L1 was found in African coelacanth. Nonetheless, the most successful LINEs in this group are still CR1 and L2 [56]. A high diversity of L1 was observed in amphibians, although, similar to that in bony fish, the number of LINEs remains small and most of them are either DNA transposons or LTR [53,54,58]. In reptiles, except green anole, several LINE families (CR1, BovB, L2) are evolutionarily successful, the activity of which continues to persist in the genomes, and their amount increases relative to other repeats [55,63,64,65,66,67,68]. The genome of tuatara is distinguished by a variety of repeats in which L2 is the most successful group [68]. The most widespread CR1, completely displacing L1, is in the genomes of turtles, crocodiles, and birds. The success of these elements was facilitated by the highly conserved hairpin structure and octameric microsatellite motif at their 3′UTR [65,69,70,71]. In the avian genomes, there was a sharp decrease in the genome size and number of repeated sequences. LINE/CR1 are the remaining bulk [71].
4.2. LINE Evolution in Mammals
4.2. LINE Evolution in Mammals
4.3. LINE Evolution in Primates
In mammals, many LINEs lost the ability to spread due to various mutations and truncations of full-length copies [64,72]. Only one family of LINEs remains active. The most successful group of mobile elements in mammals is L1. An exception is the group of monotremes, which have no L1 sequences [52]. Thus, in platypus, L2 is the most prevalent of LINEs [73]. Metatheria (marsupials) and eutheria (placental mammals) have similarities in the composition and evolutionary tendencies of their mobile elements. In the genomes of most mammalian species, L1 becomes the most successful and active group, while many ancient repeats gradually disappear from the genome in process of evolution. Interestingly, that some enhancers and ultra-conserved elements are originated from ancient retrotransposon repeats [55,74,75]. Active L1s are species-specific genomic elements. Nevertheless, their structure is similar in all mammals and the greatest differences involve the non-coding region 5′UTR, the size of which varies greatly in different species [64]. 5′UTR changes play an important role in the interaction with cellular transcription factors that regulate L1 expression. Of the L1 encoded proteins, ORF1 differs in variability, while ORF2, on the contrary, is conservative [64]. Differences and evolutionary trends of L1 elements in mammals have been described in some animal groups. For example, some bats, similar to flying birds, have a decrease size of genomes and are characterized loss of active L1 elements [71,76]. The L1 extinction is also observed in certain mammalian species that are not adapted to flight. The disappearance of L1 activity was noted for Spermophilus tridecemlineatus from the superorder Afrotheria, perissodactyls, and sigmodontine rodents [77,78,79,80].
4.4. LINE Evolution in Ancient and Modern Humans
L1 is active in rats and mice. However, the accumulation and activity of mobile genetic elements of the widely studied mouse (Mus musculus) differ from the general tendencies of mammals, including humans and other primates, because LINE elements are quantitatively inferior to LTR repeats in its genome [81]. However, L1 makes up about 20% of the murine genome and L1Md is currently active. LINEs account for about 23% of the rat genome [82]. In addition to the traditional L1, the rat genome acquired the activity of HAL1 (HALF-L1) elements, the shorter version of L1 elements. In follow up the integration into the genome the HAL1 elements retain their internal promoter, that is often truncated in case of integration of full-length L1 elements.
4.5. LINE Evolution and Host Regulation
4.3. LINE Evolution in Primates
Primates separated from other ancestral mammals about 90–65 million years (myrs) ago and are characterized by the distribution of the L1PA-L1PB families [83,84,85]. Comparative evolutionary analysis of L1 revealed different trends in discrete primate species [86]. In most primate species, L1 is the most active family capable of self-propagation in the genome, as well as the most capable of contributing to the amplification of SINE elements, the copy number of which in genomes reaches the maximum of all dispersed repeats. In most species of New and Old World primates, the L1 remains active. Only in New World South American spider monkeys the absence of L1 activity was found [87,88]. The evolutionary history of the Old World primates began approximately 21–25 myrs and is associated with the distribution of L1PA6—L1PA5 elements [83,89,90,91]. L1PA5–6 elements, which are evolutionarily closest to their modern active L1 subfamilies, are most widely distributed in the genomes of monkeys (Cercopithecoidea) [89,90]. Interestingly, the greatest differences in the number of primate L1 were found among the Cercopithecoidea. For example, the baboon has the highest L1 amplification rates in the genome compared to other primates. On the contrary, the green macaque has the lowest number of L1 repeats compared with other primates [86].
5. Regulation of L1 Activity
The branch of great apes split off about 26 myrs ago [92]. Among the great apes, the largest number of L1 insertion loci was found in the orangutan. Moreover, the number of LINEs in orangutan genome significantly dominates over other families of dispersed DNA repeats, while in other primates, SINE insertions are most common [86]. Compared to other primates, the number of LINE insertions in humans is not large. However, the largest number of currently active LINE elements was found in the human genome [86]. Thus, the gorilla genome harbors twelve intact full-length gorilla-specific L1s belonging to the L1PA2 subfamily [93]. In chimpanzees, L1Pt-2 are active and only nine copies are full-length elements with intact ORFs [94]. In contrast, in humans, the active family is the L1HS, consisting of several subfamilies, namely pre-Ta, Ta-0, Ta-1, Ta1-d, and Ta1-nd [9,95] [, of which about 146 copies are active [5]. Moreover, comparative analysis showed that the activity of human L1 copies is significantly higher than that of chimpanzees [90,94].
4.4. LINE Evolution in Ancient and Modern Humans
A number of studies have shown that the accumulation of loci containing L1 repeat insertions is not random but occurs in accordance with functional significance. Thus, L1 insertions are more often retained in the trans-orientation relative to the gene, while insertions in the cis-orientation are washed out from the genome [96,97]. The evolutionary trends of L1 in the Homo branch are of great interest. However, the genomic architecture of L1 elements in ancient humans (Homo sapiens sapiens) and related subspecies, ancient hominids (Neanderthals and Denisovans), are poorly understood due to the difficulties of genomic mapping of repeat elements using short reads available from the sequencing of ancient DNA. Nevertheless, several studies carried out an analysis of the mobile elements, which showed the presence of introgression of the L1 insertion loci of ancient people in the DNA of modern people, the nature of which corresponds to the same for SNV [98,99]. Moreover, in the genomes of ancient hominids, the sequences corresponding to the most active L1Ta1d mobile elements of the modern human genome were determined. Thus, the origin of L1Ta1d could have occurred in the common ancestor of ancient hominids and modern humans more than 800 thousand years ago [98]. An analysis of the insertion loci in genes in ancient people and modern humans showed that most of the repeat insertion loci specific to modern humans, including L1, originated in the genes that are highly expressed in the brain and are involved in neuronal maturation [99].
Analysis of L1 insertions in modern world human populations of the Phase3 data release project 1000 Genomes, which included 2.5 thousand individuals from 26 populations, reveals 2.91 thousand polymorphic L1 loci [100]. It was found that the majority (over 93%) of the identified loci of active retrotransposons (L1, Alu, and SVA) have a low population frequency of less than 5%. Moreover, such low frequency of insertion loci have substantial geographic differentiation. In support of this, in a recent study with a significantly smaller number of individuals (296 individuals) but greater population diversity (146 populations) from the Simons Genome Diversity Project (SGDP), a relatively large number of 1.886 thousand polymorphic unreferenced L1 loci were identified [101]. In both studies, the number of L1 polymorphic loci is 6–10 times lower than the Alu polymorphic loci but exceeds 3.5–4 times the number of SVA element polymorphic loci. The polymorphism of the insertion loci of active retrotransposons reflects the evolutionary aspects of modern populations and the migration processes of the world [100,101]. The greatest diversity is observed in Africans who are evolutionarily basal in world populations [100,101]. A decrease in heterozygosity is observed in populations of Eurasia and a minimum value was found in Native Americans [101].
4.5. LINE Evolution and Host Regulation
The L1 regulatory factors are evolved along with evolution of L1 elements. APOBEC3 protein family and the Piwi-interacting RNA (piRNA)-signaling pathway are involved into the cellular defense mechanisms against the uncontrolled spread of L1 (see Regulation of L1 Activity). One of the most susceptible proteins to strong evolutionary selection, amplification, and divergence in mammalian genomes is the APOBEC3 subfamily of antiviral factor genes [102,103]. High divergence of APOBEC3 was noted in the genomes of bats and primates [104,105,106]. Interestingly, other closely related genes belonging to the AID/APOBEC family have lower evolutionary rates in mammals [107]. High evolutionary rates are also observed for the piRNA pathway, many genes of which are under the influence of positive selection [108]. The different regulatory pathways capable to repress L1 elements have been evolved reflecting the constant battle between mobile elements and the cellular host defence [109,110,111]. The difference in expression of genes involved in host defence pathways of mobile elements between animal species plays an important role in the effectiveness of L1 inhibition. As shown in one study, there is a higher expression level of APOBEC3B (also known as A3B) and PIWIL2 genes in human pluripotent stem cells, compared to the closest non-human primates (Pan troglodytes and Pan paniscus). The study showed that L1 silencing in human cells is more efficient as compared to chimpanzee cells [112].
Further, the factors regulating L1 elements are considered in detail.
- Regulation of L1 Activity
The process of the regulation of L1 activity throughout ontogenesis is complicated. In most cells, L1 activity is inhibited at all stages of the retrotransposition process in various ways: by decreasing the availability of DNA using DNA methylation [110,113,114], histone modifications, and heterochromatin formation [110,115,116]; through post-transcriptional inhibition by degradation of new RNA copies of L1 [117,118]; through repression of ORF1 and ORF2 translation; through the binding of L1 RNPs and the obstruction of their transportation to the nucleus [119,120,121,122]; and, at the last stage of integration for a new copy of the L1 element into the genome, through using DNA repair mechanisms [120,123,124,125,126] (
Figure 3). In the process of organism ontogenesis, changes in the regulation of L1 activity occur. Thus, at the stage of formation of germ cells and mature germ cells, L1 poses a great threat to the future organism and, therefore, is thoroughly repressed by cells [127,128]. Most experimental knockouts of factors involved in the L1-silencing in germ cells lead to their death and infertility [129]. In the early stages of embryogenesis, activity of L1 is also dangerous for the developing organism and, therefore, is repressed [130,131,132]. Some changes occur in the pathways of L1 repression during embryogenesis and the activity of L1 elements increases at certain stages [133,134]. L1 elements are mainly repressed in somatic tissues of a mature organism, but increased L1 activity is noted for some pathologies including cancer as well as autoimmune and neuropsychiatric disorders [135]. With normal ageing, changes in the number of insertions of L1 are insignificant [136]. However, in some tissues, especially in the brain, L1 is not completely suppressed and L1 retrotranspositions can be activated [137,138,139].
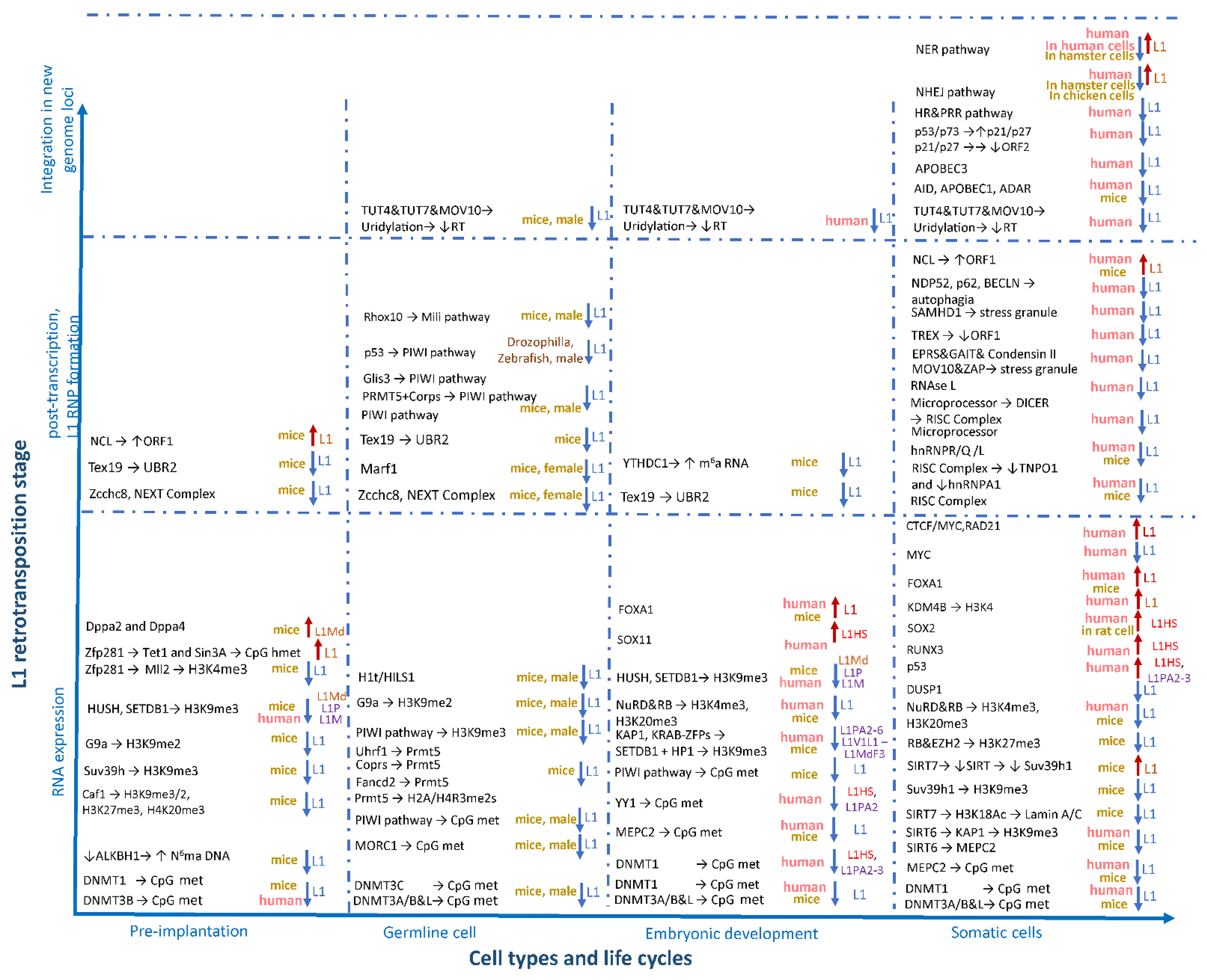
Figure 3. Factors affecting the activity of L1 retrotransposons during ontogenesis. The factors involved in L1 regulation are grouped horizontally in accordance with the stages of prenatal development (pre- and post-implantation period and in germline cells) and the postnatal period (somatic cells), as well as vertically depending on the stage of the retrotransposition process (expression, L1 RNP formation, and integration into the genome).
- Factors Affecting Changes in L1 Regulation in Neuropsychiatric Diseases
Normally, L1 can be active in the brain [113,138,139,234,235,236]. In some neuropsychological pathologies, changes in L1 activity were detected. The most pronounced increase in L1 activity was found in Rett syndrome [241] and autism [400], as well as in ataxia telangiectasia [45]. The genetic causes that lead to an increase in L1 activity have been mostly studied with Rett syndrome and ataxia telangiectasia, and are associated with damage in the MEPC2 and ATM genes [45,113]. The L1 control mechanism of these genes is described above. Some trends are observed in schizophrenia [401,402] and major depressive disorder [403]. However, the causes and factors that change the activity of L1 elements remain unknown for most diseases. Recently, some studies demonstrated the connection of genetic factors associated with neurodegenerative pathologies and L1 activity. One of these factors is the TAR DNA-binding protein (TDP-43), which is able to bind DNA and RNA, and is involved in the regulation of many processes [404]. TDP-43 is associated with neuropsychiatric pathologies such as amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD) [405]. The protein cleavage, hyperphosphorylation, and aggregation in the form of ubiquitinated granules in the cytoplasm occur in the pathologies. Similar the TDP-43 “proteinopathy” occurs in other neurodegenerative diseases such as Alzheimer’s disease [406], Parkinson’s disease [407], and Huntington’s disease [408], and also with hereditary inclusion body myopathy (HIBM) [409]. Controversial results have been obtained regarding the effect of TDP-43 on L1 activity. TDP-43 is involved in many processes that can affect L1 activity, such as in autophagy, which contributes to the destruction of L1 stress granules [410], and in double-stranded DNA repair, wherein it binds to the damaged site and provides further formation of the XRCC4-DNA ligase IV complex, the activity of which can contribute to retrotransposition [49,50,411]. Additionally, in one of the latest studies, data were obtained regarding the inhibitory effect of TDP-43 on L1 activity and its absence was found to increase the level of L1 retrotranspositions by chromatin decompactivation [412]. Despite this, other studies obtained different results, showing that TDP-43 regulates the transcription of many genes and retrotransposons of Alu elements, and does not affect the activity of L1 elements [413,414]. In addition, an increase in HERV-K retroviral repeats was noted, while no changes in L1 activity were detected in lateral amyotrophic sclerosis [415,416]. The change in the expression of retrotransposons is also associated with the Tau protein encoded by the MAPT (microtubule-associated protein tau) gene [417,418]. Tau pathology is observed in various neurodegenerative disorders including Alzheimer’s disease [419,420]. The Tau protein hyperphosphorylates and forms hyperphosphorylated insoluble aggregates called neurofibrillary tangles [421,422,423]. One study showed activation of various retrotransposons, including L1 that lost the ability to retrotranspose due to accumulated mutations in the transcriptional reading frame [418]. We revealed no significant changes in the copy number of L1 in Alzheimer’s disease [136]. Another study showed an increase in the expression of endogenous retroviruses, but not of active L1, as a result of chromatin decondensation and a decrease in both piRNA and piwi proteins associated with Tau pathology in Alzheimer’s disease [417]. Mitochondrial dysfunction and oxidative stress are characteristic features of a number of diseases such as some forms of ataxia, neurodegenerative diseases (Parkinson’s disease in particular), various forms of cancer, and other diseases [424,425,426,427]. Recent studies have shown that abnormalities and a deficiency of both the mitochondrial chain and oxidative stress cause DNA hypomethylation and increased L1 activity [428,429,430]. The stress sensor GABB45B gene was connected to the death of dopaminergic neurons in Parkinson’s disease [431]. A recent study in mice shows that overexpression of Gadd45b leads to disorganized heterochromatin, increased DNA damage, vulnerability to oxidative stress, and further stable changes in DNA methylation, particularly in introns of neuronal genes harboring L1 [432].
6. Factors Affecting Changes in L1 Regulation in Neuropsychiatric Diseases
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921.
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which Transposable Elements Are Active in the Human Genome? Trends Genet. 2007, 23, 183–191.
- Sassaman, D.M.; Dombroski, B.A.; Moran, J.V.; Kimberland, M.L.; Naas, T.P.; DeBerardinis, R.J.; Gabriel, A.; Swergold, G.D.; Kazazian, H.H. Many Human L1 Elements Are Capable of Retrotransposition. Nat. Genet. 1997, 16, 37–43.
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H. Hot L1s Account for the Bulk of Retrotransposition in the Human Population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285.
- Penzkofer, T.; Jäger, M.; Figlerowicz, M.; Badge, R.; Mundlos, S.; Robinson, P.N.; Zemojtel, T. L1Base 2: More Retrotransposition-Active LINE-1s, More Mammalian Genomes. Nucleic Acids Res. 2017, 45, D68–D73.
- Dewannieux, M.; Esnault, C.; Heidmann, T. LINE-Mediated Retrotransposition of Marked Alu Sequences. Nat. Genet. 2003, 35, 41–48.
- Ostertag, E.M.; Goodier, J.L.; Zhang, Y.; Kazazian, H.H. SVA Elements Are Nonautonomous Retrotransposons That Cause Disease in Humans. Am. J. Hum. Genet. 2003, 73, 1444–1451.
- Wang, H.; Xing, J.; Grover, D.; Hedges, D.J.; Han, K.; Walker, J.A.; Batzer, M.A. SVA Elements: A Hominid-Specific Retroposon Family. J. Mol. Biol. 2005, 354, 994–1007.
- Xing, J.; Zhang, Y.; Han, K.; Salem, A.H.; Sen, S.K.; Huff, C.D.; Zhou, Q.; Kirkness, E.F.; Levy, S.; Batzer, M.A.; et al. Mobile Elements Create Structural Variation: Analysis of a Complete Human Genome. Genome Res. 2009, 19, 1516–1526.
- Mandal, P.K.; Ewing, A.D.; Hancks, D.C.; Kazazian, H.H. Enrichment of Processed Pseudogene Transcripts in L1-Ribonucleoprotein Particles. Hum. Mol. Genet. 2013, 22, 3730–3748.
- Pavlícek, A.; Paces, J.; Zíka, R.; Hejnar, J. Length Distribution of Long Interspersed Nucleotide Elements (LINEs) and Processed Pseudogenes of Human Endogenous Retroviruses: Implications for Retrotransposition and Pseudogene Detection. Gene 2002, 300, 189–194.
- Scott, A.F.; Schmeckpeper, B.J.; Abdelrazik, M.; Comey, C.T.; O’Hara, B.; Rossiter, J.P.; Cooley, T.; Heath, P.; Smith, K.D.; Margolet, L. Origin of the Human L1 Elements: Proposed Progenitor Genes Deduced from a Consensus DNA Sequence. Genomics 1987, 1, 113–125.
- Mätlik, K.; Redik, K.; Speek, M. L1 Antisense Promoter Drives Tissue-Specific Transcription of Human Genes. J. Biomed. Biotechnol. 2006, 2006, 71753.
- Swergold, G.D. Identification, Characterization, and Cell Specificity of a Human LINE-1 Promoter. Mol. Cell. Biol. 1990, 10, 6718–6729.
- Moran, J.V.; Holmes, S.E.; Naas, T.P.; DeBerardinis, R.J.; Boeke, J.D.; Kazazian, H.H. High Frequency Retrotransposition in Cultured Mammalian Cells. Cell 1996, 87, 917–927.
- Kolosha, V.O.; Martin, S.L. In Vitro Properties of the First ORF Protein from Mouse LINE-1 Support Its Role in Ribonucleoprotein Particle Formation during Retrotransposition. Proc. Natl. Acad. Sci. USA 1997, 94, 10155–10160.
- Naufer, M.N.; Furano, A.V.; Williams, M.C. Protein-Nucleic Acid Interactions of LINE-1 ORF1p. Semin. Cell Dev. Biol. 2019, 86, 140–149.
- Freeman, B.T.; Sokolowski, M.; Roy-Engel, A.M.; Smither, M.E.; Belancio, V.P. Identification of Charged Amino Acids Required for Nuclear Localization of Human L1 ORF1 Protein. Mob. DNA 2019, 10, 20.
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H.; Boeke, J.D.; Gabriel, A. Reverse Transcriptase Encoded by a Human Transposable Element. Science 1991, 254, 1808–1810.
- Feng, Q.; Moran, J.V.; Kazazian, H.H.; Boeke, J.D. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 1996, 87, 905–916.
- Macia, A.; Muñoz-Lopez, M.; Cortes, J.L.; Hastings, R.K.; Morell, S.; Lucena-Aguilar, G.; Marchal, J.A.; Badge, R.M.; Garcia-Perez, J.L. Epigenetic Control of Retrotransposon Expression in Human Embryonic Stem Cells. Mol. Cell. Biol. 2011, 31, 300–316.
- Wheelan, S.J.; Aizawa, Y.; Han, J.S.; Boeke, J.D. Gene-Breaking: A New Paradigm for Human Retrotransposon-Mediated Gene Evolution. Genome Res. 2005, 15, 1073–1078.
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.N.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 2015, 163, 583–593.
- Adney, E.M.; Ochmann, M.T.; Sil, S.; Truong, D.M.; Mita, P.; Wang, X.; Kahler, D.J.; Fenyö, D.; Holt, L.J.; Boeke, J.D. Comprehensive Scanning Mutagenesis of Human Retrotransposon LINE-1 Identifies Motifs Essential for Function. Genetics 2019, 213, 1401–1414.
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse Transcription of R2Bm RNA Is Primed by a Nick at the Chromosomal Target Site: A Mechanism for Non-LTR Retrotransposition. Cell 1993, 72, 595–605.
- Cost, G.J.; Feng, Q.; Jacquier, A.; Boeke, J.D. Human L1 Element Target-Primed Reverse Transcription in Vitro. EMBO J. 2002, 21, 5899–5910.
- Athanikar, J.N.; Badge, R.M.; Moran, J.V. A YY1-Binding Site Is Required for Accurate Human LINE-1 Transcription Initiation. Nucleic Acids Res. 2004, 32, 3846–3855.
- Dai, L.; Taylor, M.S.; O’Donnell, K.A.; Boeke, J.D. Poly(A) Binding Protein C1 Is Essential for Efficient L1 Retrotransposition and Affects L1 RNP Formation. Mol. Cell. Biol. 2012, 32, 4323–4336.
- Doucet, A.J.; Hulme, A.E.; Sahinovic, E.; Kulpa, D.A.; Moldovan, J.B.; Kopera, H.C.; Athanikar, J.N.; Hasnaoui, M.; Bucheton, A.; Moran, J.V.; et al. Characterization of LINE-1 Ribonucleoprotein Particles. PLoS Genet. 2010, 6, e1001150.
- Horn, A.V.; Celic, I.; Dong, C.; Martirosyan, I.; Han, J.S. A Conserved Role for the ESCRT Membrane Budding Complex in LINE Retrotransposition. PLoS Genet. 2017, 13, e1006837.
- Mita, P.; Wudzinska, A.; Sun, X.; Andrade, J.; Nayak, S.; Kahler, D.J.; Badri, S.; LaCava, J.; Ueberheide, B.; Yun, C.Y.; et al. LINE-1 Protein Localization and Functional Dynamics during the Cell Cycle. eLife 2018, 7, e30058.
- Macia, A.; Widmann, T.J.; Heras, S.R.; Ayllon, V.; Sanchez, L.; Benkaddour-Boumzaouad, M.; Muñoz-Lopez, M.; Rubio, A.; Amador-Cubero, S.; Blanco-Jimenez, E.; et al. Engineered LINE-1 Retrotransposition in Nondividing Human Neurons. Genome Res. 2017, 27, 335–348.
- Cost, G.J.; Boeke, J.D. Targeting of Human Retrotransposon Integration Is Directed by the Specificity of the L1 Endonuclease for Regions of Unusual DNA Structure. Biochemistry 1998, 37, 18081–18093.
- Flasch, D.A.; Macia, Á.; Sánchez, L.; Ljungman, M.; Heras, S.R.; García-Pérez, J.L.; Wilson, T.E.; Moran, J.V. Genome-Wide de Novo L1 Retrotransposition Connects Endonuclease Activity with Replication. Cell 2019, 177, 837–851.e28.
- Sultana, T.; van Essen, D.; Siol, O.; Bailly-Bechet, M.; Philippe, C.; Zine El Aabidine, A.; Pioger, L.; Nigumann, P.; Saccani, S.; Andrau, J.-C.; et al. The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-Insertion Sequence Biases and Post-Insertion Selection. Mol. Cell 2019, 74, 555–570.e7.
- Doucet, A.J.; Wilusz, J.E.; Miyoshi, T.; Liu, Y.; Moran, J.V. A 3′ Poly(A) Tract Is Required for LINE-1 Retrotransposition. Mol. Cell 2015, 60, 728–741.
- Kulpa, D.A.; Moran, J.V. Cis-Preferential LINE-1 Reverse Transcriptase Activity in Ribonucleoprotein Particles. Nat. Struct. Mol. Biol. 2006, 13, 655–660.
- Monot, C.; Kuciak, M.; Viollet, S.; Mir, A.A.; Gabus, C.; Darlix, J.-L.; Cristofari, G. The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts. PLoS Genet. 2013, 9, e1003499.
- Gibson, B.A.; Kraus, W.L. New Insights into the Molecular and Cellular Functions of Poly(ADP-Ribose) and PARPs. Nat. Rev. Mol. Cell. Biol. 2012, 13, 411–424.
- Miyoshi, T.; Makino, T.; Moran, J.V. Poly(ADP-Ribose) Polymerase 2 Recruits Replication Protein A to Sites of LINE-1 Integration to Facilitate Retrotransposition. Mol. Cell 2019, 75, 1286–1298.e12.
- Chen, R.; Wold, M.S. Replication Protein A: Single-Stranded DNA’s First Responder: Dynamic DNA-Interactions Allow Replication Protein A to Direct Single-Strand DNA Intermediates into Different Pathways for Synthesis or Repair. Bioessays 2014, 36, 1156–1161.
- Taylor, M.S.; Altukhov, I.; Molloy, K.R.; Mita, P.; Jiang, H.; Adney, E.M.; Wudzinska, A.; Badri, S.; Ischenko, D.; Eng, G.; et al. Dissection of Affinity Captured LINE-1 Macromolecular Complexes. eLife 2018, 7, e30094.
- Taylor, M.S.; LaCava, J.; Mita, P.; Molloy, K.R.; Huang, C.R.L.; Li, D.; Adney, E.M.; Jiang, H.; Burns, K.H.; Chait, B.T.; et al. Affinity Proteomics Reveals Human Host Factors Implicated in Discrete Stages of LINE-1 Retrotransposition. Cell 2013, 155, 1034–1048.
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 Is a 5′ to 3′ RNA Helicase Contributing to UPF1 MRNA Target Degradation by Translocation along 3′ UTRs. Mol. Cell 2014, 54, 573–585.
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Marchetto, M.C.N.; Muotri, A.R.; Mu, Y.; Carson, C.T.; Macia, A.; Moran, J.V.; Gage, F.H. Ataxia Telangiectasia Mutated (ATM) Modulates Long Interspersed Element-1 (L1) Retrotransposition in Human Neural Stem Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20382–20387.
- Suzuki, J.; Yamaguchi, K.; Kajikawa, M.; Ichiyanagi, K.; Adachi, N.; Koyama, H.; Takeda, S.; Okada, N. Genetic Evidence That the Non-Homologous End-Joining Repair Pathway Is Involved in LINE Retrotransposition. PLoS Genet. 2009, 5, e1000461.
- Zingler, N.; Willhoeft, U.; Brose, H.-P.; Schoder, V.; Jahns, T.; Hanschmann, K.-M.O.; Morrish, T.A.; Löwer, J.; Schumann, G.G. Analysis of 5′ Junctions of Human LINE-1 and Alu Retrotransposons Suggests an Alternative Model for 5′-End Attachment Requiring Microhomology-Mediated End-Joining. Genome Res. 2005, 15, 780–789.
- Kopera, H.C.; Moldovan, J.B.; Morrish, T.A.; Garcia-Perez, J.L.; Moran, J.V. Similarities between Long Interspersed Element-1 (LINE-1) Reverse Transcriptase and Telomerase. Proc. Natl. Acad. Sci. USA 2011, 108, 20345–20350.
- Morrish, T.A.; Garcia-Perez, J.L.; Stamato, T.D.; Taccioli, G.E.; Sekiguchi, J.; Moran, J.V. Endonuclease-Independent LINE-1 Retrotransposition at Mammalian Telomeres. Nature 2007, 446, 208–212.
- Morrish, T.A.; Gilbert, N.; Myers, J.S.; Vincent, B.J.; Stamato, T.D.; Taccioli, G.E.; Batzer, M.A.; Moran, J.V. DNA Repair Mediated by Endonuclease-Independent LINE-1 Retrotransposition. Nat. Genet. 2002, 31, 159–165.
- Ohshima, K. Parallel Relaxation of Stringent RNA Recognition in Plant and Mammalian L1 Retrotransposons. Mol. Biol. Evol. 2012, 29, 3255–3259.
- Ivancevic, A.M.; Kortschak, R.D.; Bertozzi, T.; Adelson, D.L. LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life. Genome Biol. Evol. 2016, 8, 3301–3322.
- Kordis, D.; Lovsin, N.; Gubensek, F. Phylogenomic Analysis of the L1 Retrotransposons in Deuterostomia. Syst. Biol. 2006, 55, 886–901.
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.-N. Comparative Analysis of Transposable Elements Highlights Mobilome Diversity and Evolution in Vertebrates. Genome Biol. Evol. 2015, 7, 567–580.
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and Diversity of Transposable Elements in Vertebrate Genomes. Genome Biol. Evol. 2017, 9, 161–177.
- Shao, F.; Han, M.; Peng, Z. Evolution and Diversity of Transposable Elements in Fish Genomes. Sci. Rep. 2019, 9, 15399.
- Metcalfe, C.J.; Filée, J.; Germon, I.; Joss, J.; Casane, D. Evolution of the Australian Lungfish (Neoceratodus Forsteri) Genome: A Major Role for CR1 and L2 LINE Elements. Mol. Biol. Evol. 2012, 29, 3529–3539.
- Nikaido, M.; Noguchi, H.; Nishihara, H.; Toyoda, A.; Suzuki, Y.; Kajitani, R.; Suzuki, H.; Okuno, M.; Aibara, M.; Ngatunga, B.P.; et al. Coelacanth Genomes Reveal Signatures for Evolutionary Transition from Water to Land. Genome Res. 2013, 23, 1740–1748.
- Luchetti, A.; Plazzi, F.; Mantovani, B. Evolution of Two Short Interspersed Elements in Callorhinchus milii (Chondrichthyes, Holocephali) and Related Elements in Sharks and the Coelacanth. Genome Biol. Evol. 2017, 9, 1406–1417.
- Meyer, A.; Schloissnig, S.; Franchini, P.; Du, K.; Woltering, J.M.; Irisarri, I.; Wong, W.Y.; Nowoshilow, S.; Kneitz, S.; Kawaguchi, A.; et al. Giant Lungfish Genome Elucidates the Conquest of Land by Vertebrates. Nature 2021, 590, 284–289.
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant Shark Genome Provides Unique Insights into Gnathostome Evolution. Nature 2014, 505, 174–179.
- Smith, J.J.; Kuraku, S.; Holt, C.; Sauka-Spengler, T.; Jiang, N.; Campbell, M.S.; Yandell, M.D.; Manousaki, T.; Meyer, A.; Bloom, O.E.; et al. Sequencing of the Sea Lamprey (Petromyzon Marinus) Genome Provides Insights into Vertebrate Evolution. Nat. Genet. 2013, 45, 415–421.
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The Genome of the Green Anole Lizard and a Comparative Analysis with Birds and Mammals. Nature 2011, 477, 587–591.
- Boissinot, S.; Sookdeo, A. The Evolution of LINE-1 in Vertebrates. Genome Biol. Evol. 2016, 8, 3485–3507.
- Suh, A.; Churakov, G.; Ramakodi, M.P.; Platt, R.N.; Jurka, J.; Kojima, K.K.; Caballero, J.; Smit, A.F.; Vliet, K.A.; Hoffmann, F.G.; et al. Multiple Lineages of Ancient CR1 Retroposons Shaped the Early Genome Evolution of Amniotes. Genome Biol. Evol. 2014, 7, 205–217.
- Pasquesi, G.I.M.; Adams, R.H.; Card, D.C.; Schield, D.R.; Corbin, A.B.; Perry, B.W.; Reyes-Velasco, J.; Ruggiero, R.P.; Vandewege, M.W.; Shortt, J.A.; et al. Squamate Reptiles Challenge Paradigms of Genomic Repeat Element Evolution Set by Birds and Mammals. Nat. Commun. 2018, 9, 2774.
- Boissinot, S.; Bourgeois, Y.; Manthey, J.D.; Ruggiero, R.P. The Mobilome of Reptiles: Evolution, Structure, and Function. Cytogenet. Genome Res. 2019, 157, 21–33.
- Gemmell, N.J.; Rutherford, K.; Prost, S.; Tollis, M.; Winter, D.; Macey, J.R.; Adelson, D.L.; Suh, A.; Bertozzi, T.; Grau, J.H.; et al. The Tuatara Genome Reveals Ancient Features of Amniote Evolution. Nature 2020, 584, 403–409.
- Shedlock, A.M. Phylogenomic Investigation of CR1 LINE Diversity in Reptiles. Syst. Biol. 2006, 55, 902–911.
- Suh, A. The Specific Requirements for CR1 Retrotransposition Explain the Scarcity of Retrogenes in Birds. J. Mol. Evol. 2015, 81, 18–20.
- Kapusta, A.; Suh, A. Evolution of Bird Genomes-a Transposon’s-Eye View. Ann. N. Y. Acad. Sci. 2017, 1389, 164–185.
- Smit, A.F.; Tóth, G.; Riggs, A.D.; Jurka, J. Ancestral, Mammalian-Wide Subfamilies of LINE-1 Repetitive Sequences. J. Mol. Biol. 1995, 246, 401–417.
- Warren, W.C.; Hillier, L.W.; Marshall Graves, J.A.; Birney, E.; Ponting, C.P.; Grützner, F.; Belov, K.; Miller, W.; Clarke, L.; Chinwalla, A.T.; et al. Genome Analysis of the Platypus Reveals Unique Signatures of Evolution. Nature 2008, 453, 175–183.
- Mikkelsen, T.S.; Wakefield, M.J.; Aken, B.; Amemiya, C.T.; Chang, J.L.; Duke, S.; Garber, M.; Gentles, A.J.; Goodstadt, L.; Heger, A.; et al. Genome of the Marsupial Monodelphis Domestica Reveals Innovation in Non-Coding Sequences. Nature 2007, 447, 167–177.
- Polychronopoulos, D.; King, J.W.D.; Nash, A.J.; Tan, G.; Lenhard, B. Conserved Non-Coding Elements: Developmental Gene Regulation Meets Genome Organization. Nucleic Acids Res. 2017, 45, 12611–12624.
- Wichman, H.A.; Scott, L.; Howell, E.K.; Martinez, A.R.; Yang, L.; Baker, R.J. Flying Around in the Genome: Characterization of LINE-1 in Chiroptera. Spec. Publ. Tex. Tech. Univ. Mus. 2019, 71, 379–392.
- Platt, R.N.; Ray, D.A. A Non-LTR Retroelement Extinction in Spermophilus Tridecemlineatus. Gene 2012, 500, 47–53.
- Sookdeo, A.; Hepp, C.M.; Boissinot, S. Contrasted Patterns of Evolution of the LINE-1 Retrotransposon in Perissodactyls: The History of a LINE-1 Extinction. Mob. DNA 2018, 9, 12.
- Yang, L.; Scott, L.; Wichman, H.A. Tracing the History of LINE and SINE Extinction in Sigmodontine Rodents. Mob. DNA 2019, 10, 22.
- Blumenstiel, J.P. Birth, School, Work, Death, and Resurrection: The Life Stages and Dynamics of Transposable Element Proliferation. Genes 2019, 10, 336.
- Mouse Genome Sequencing Consortium; Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; et al. Initial Sequencing and Comparative Analysis of the Mouse Genome. Nature 2002, 420, 520–562.
- Gibbs, R.A.; Weinstock, G.M.; Metzker, M.L.; Muzny, D.M.; Sodergren, E.J.; Scherer, S.; Scott, G.; Steffen, D.; Worley, K.C.; Burch, P.E.; et al. Genome Sequence of the Brown Norway Rat Yields Insights into Mammalian Evolution. Nature 2004, 428, 493–521.
- Glazko, G.V.; Nei, M. Estimation of Divergence Times for Major Lineages of Primate Species. Mol. Biol. Evol. 2003, 20, 424–434.
- Chatterjee, H.J.; Ho, S.Y.W.; Barnes, I.; Groves, C. Estimating the Phylogeny and Divergence Times of Primates Using a Supermatrix Approach. BMC Evol. Biol. 2009, 9, 259.
- Konkel, M.K.; Walker, J.A.; Batzer, M.A. LINEs and SINEs of Primate Evolution. Evol. Anthropol. 2010, 19, 236–249.
- Tang, W.; Liang, P. Comparative Genomics Analysis Reveals High Levels of Differential Retrotransposition among Primates from the Hominidae and the Cercopithecidae Families. Genome Biol. Evol. 2019, 11, 3309–3325.
- Boissinot, S.; Roos, C.; Furano, A.V. Different Rates of LINE-1 (L1) Retrotransposon Amplification and Evolution in New World Monkeys. J. Mol. Evol. 2004, 58, 122–130.
- Sookdeo, A.; Ruiz-García, M.; Schneider, H.; Boissinot, S. Contrasting Rates of LINE-1 Amplification among New World Primates of the Atelidae Family. Cytogenet. Genome Res. 2018, 154, 217–228.
- Han, K.; Konkel, M.K.; Xing, J.; Wang, H.; Lee, J.; Meyer, T.J.; Huang, C.T.; Sandifer, E.; Hebert, K.; Barnes, E.W.; et al. Mobile DNA in Old World Monkeys: A Glimpse through the Rhesus Macaque Genome. Science 2007, 316, 238–240.
- Lee, S.; Tang, W.; Liang, P.; Han, K. A Comprehensive Analysis of Chimpanzee (Pan Troglodytes)-Specific LINE-1 Retrotransposons. Gene 2019, 693, 46–51.
- Fernandes, J.D.; Zamudio-Hurtado, A.; Clawson, H.; Kent, W.J.; Haussler, D.; Salama, S.R.; Haeussler, M. The UCSC Repeat Browser Allows Discovery and Visualization of Evolutionary Conflict across Repeat Families. Mob. DNA 2020, 11, 13.
- Castellano, D.; Munch, K. Population Genomics in the Great Apes. Methods Mol. Biol. 2020, 2090, 453–463.
- Jeon, S.; Kim, S.; Oh, M.H.; Liang, P.; Tang, W.; Han, K. A Comprehensive Analysis of Gorilla-Specific LINE-1 Retrotransposons. Genes Genom. 2021, 43, 1133–1141.
- Lee, J.; Cordaux, R.; Han, K.; Wang, J.; Hedges, D.J.; Liang, P.; Batzer, M.A. Different Evolutionary Fates of Recently Integrated Human and Chimpanzee LINE-1 Retrotransposons. Gene 2007, 390, 18–27.
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 Retrotransposition Activity in Human Genomes. Cell 2010, 141, 1159–1170.
- Cordaux, R.; Batzer, M.A. The Impact of Retrotransposons on Human Genome Evolution. Nat. Rev. Genet. 2009, 10, 691–703.
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, MDNA3-0061-2014.
- Gardner, E.J.; Lam, V.K.; Harris, D.N.; Chuang, N.T.; Scott, E.C.; Pittard, W.S.; Mills, R.E.; 1000 Genomes Project Consortium; Devine, S.E. The Mobile Element Locator Tool (MELT): Population-Scale Mobile Element Discovery and Biology. Genome Res. 2017, 27, 1916–1929.
- Guichard, E.; Peona, V.; Malagoli Tagliazucchi, G.; Abitante, L.; Jagoda, E.; Musella, M.; Ricci, M.; Rubio-Roldán, A.; Sarno, S.; Luiselli, D.; et al. Impact of Non-LTR Retrotransposons in the Differentiation and Evolution of Anatomically Modern Humans. Mob. DNA 2018, 9, 28.
- Rishishwar, L.; Tellez Villa, C.E.; Jordan, I.K. Transposable Element Polymorphisms Recapitulate Human Evolution. Mob. DNA 2015, 6, 21.
- Watkins, W.S.; Feusier, J.E.; Thomas, J.; Goubert, C.; Mallick, S.; Jorde, L.B. The Simons Genome Diversity Project: A Global Analysis of Mobile Element Diversity. Genome Biol. Evol. 2020, 12, 779–794.
- Ito, J.; Gifford, R.J.; Sato, K. Retroviruses Drive the Rapid Evolution of Mammalian APOBEC3 Genes. Proc. Natl. Acad. Sci. USA 2020, 117, 610–618.
- Uriu, K.; Kosugi, Y.; Ito, J.; Sato, K. The Battle between Retroviruses and APOBEC3 Genes: Its Past and Present. Viruses 2021, 13, 124.
- Hayward, J.A.; Tachedjian, M.; Cui, J.; Cheng, A.Z.; Johnson, A.; Baker, M.L.; Harris, R.S.; Wang, L.-F.; Tachedjian, G. Differential Evolution of Antiretroviral Restriction Factors in Pteropid Bats as Revealed by APOBEC3 Gene Complexity. Mol. Biol. Evol. 2018, 35, 1626–1637.
- Yang, L.; Emerman, M.; Malik, H.S.; McLaughlin, R.N. Retrocopying Expands the Functional Repertoire of APOBEC3 Antiviral Proteins in Primates. eLife 2020, 9, e58436.
- Uriu, K.; Kosugi, Y.; Suzuki, N.; Ito, J.; Sato, K. Elucidation of the Complicated Scenario of Primate APOBEC3 Gene Evolution. J. Virol. 2021, 95, e00144-21.
- Harris, R.S.; Dudley, J.P. APOBECs and Virus Restriction. Virology 2015, 479–480, 131–145.
- Parhad, S.S.; Theurkauf, W.E. Rapid Evolution and Conserved Function of the PiRNA Pathway. Open Biol. 2019, 9, 180181.
- Castro-Diaz, N.; Ecco, G.; Coluccio, A.; Kapopoulou, A.; Yazdanpanah, B.; Friedli, M.; Duc, J.; Jang, S.M.; Turelli, P.; Trono, D. Evolutionally Dynamic L1 Regulation in Embryonic Stem Cells. Genes Dev. 2014, 28, 1397–1409.
- Sanchez-Luque, F.J.; Kempen, M.-J.H.C.; Gerdes, P.; Vargas-Landin, D.B.; Richardson, S.R.; Troskie, R.-L.; Jesuadian, J.S.; Cheetham, S.W.; Carreira, P.E.; Salvador-Palomeque, C.; et al. LINE-1 Evasion of Epigenetic Repression in Humans. Mol. Cell 2019, 75, 590–604.e12.
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR Is a Pseudo-PARP That Directs HUSH Complex Assembly and Epigenetic Transposon Control. Nat. Commun. 2020, 11, 4940.
- Marchetto, M.C.N.; Narvaiza, I.; Denli, A.M.; Benner, C.; Lazzarini, T.A.; Nathanson, J.L.; Paquola, A.C.M.; Desai, K.N.; Herai, R.H.; Weitzman, M.D.; et al. Differential L1 Regulation in Pluripotent Stem Cells of Humans and Apes. Nature 2013, 503, 525–529.
- Muotri, A.R.; Chu, V.T.; Marchetto, M.C.N.; Deng, W.; Moran, J.V.; Gage, F.H. Somatic Mosaicism in Neuronal Precursor Cells Mediated by L1 Retrotransposition. Nature 2005, 435, 903–910.
- Liao, J.; Karnik, R.; Gu, H.; Ziller, M.J.; Clement, K.; Tsankov, A.M.; Akopian, V.; Gifford, C.A.; Donaghey, J.; Galonska, C.; et al. Targeted Disruption of DNMT1, DNMT3A and DNMT3B in Human Embryonic Stem Cells. Nat. Genet. 2015, 47, 469–478.
- Hatanaka, Y.; Inoue, K.; Oikawa, M.; Kamimura, S.; Ogonuki, N.; Kodama, E.N.; Ohkawa, Y.; Tsukada, Y.; Ogura, A. Histone Chaperone CAF-1 Mediates Repressive Histone Modifications to Protect Preimplantation Mouse Embryos from Endogenous Retrotransposons. Proc. Natl. Acad. Sci. USA 2015, 112, 14641–14646.
- He, J.; Fu, X.; Zhang, M.; He, F.; Li, W.; Abdul, M.M.; Zhou, J.; Sun, L.; Chang, C.; Li, Y.; et al. Transposable Elements Are Regulated by Context-Specific Patterns of Chromatin Marks in Mouse Embryonic Stem Cells. Nat. Commun. 2019, 10, 34.
- Silverman, R.H. Viral Encounters with 2′,5′-Oligoadenylate Synthetase and RNase L during the Interferon Antiviral Response. J. Virol. 2007, 81, 12720–12729.
- Yao, Q.; Cao, G.; Li, M.; Wu, B.; Zhang, X.; Zhang, T.; Guo, J.; Yin, H.; Shi, L.; Chen, J.; et al. Ribonuclease Activity of MARF1 Controls Oocyte RNA Homeostasis and Genome Integrity in Mice. Proc. Natl. Acad. Sci. USA 2018, 115, 11250–11255.
- Orecchini, E.; Frassinelli, L.; Galardi, S.; Ciafrè, S.A.; Michienzi, A. Post-Transcriptional Regulation of LINE-1 Retrotransposition by AID/APOBEC and ADAR Deaminases. Chromosome Res. 2018, 26, 45–59.
- Goodier, J.L.; Pereira, G.C.; Cheung, L.E.; Rose, R.J.; Kazazian, H.H. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet. 2015, 11, e1005252.
- Herrmann, A.; Wittmann, S.; Thomas, D.; Shepard, C.N.; Kim, B.; Ferreirós, N.; Gramberg, T. The SAMHD1-Mediated Block of LINE-1 Retroelements Is Regulated by Phosphorylation. Mob. DNA 2018, 9, 11.
- Guo, L.; Byun, H.-M.; Zhong, J.; Motta, V.; Barupal, J.; Zheng, Y.; Dou, C.; Zhang, F.; McCracken, J.P.; Diaz, A.; et al. Effects of Short-Term Exposure to Inhalable Particulate Matter on DNA Methylation of Tandem Repeats. Environ. Mol. Mutagen. 2014, 55, 322–335.
- Gasior, S.L.; Roy-Engel, A.M.; Deininger, P.L. ERCC1/XPF Limits L1 Retrotransposition. DNA Repair. (Amst.) 2008, 7, 983–989.
- Servant, G.; Streva, V.A.; Derbes, R.S.; Wijetunge, M.I.; Neeland, M.; White, T.B.; Belancio, V.P.; Roy-Engel, A.M.; Deininger, P.L. The Nucleotide Excision Repair Pathway Limits L1 Retrotransposition. Genetics 2017, 205, 139–153.
- Pizarro, J.G.; Cristofari, G. Post-Transcriptional Control of LINE-1 Retrotransposition by Cellular Host Factors in Somatic Cells. Front. Cell Dev. Biol. 2016, 4, 14.
- Mita, P.; Sun, X.; Fenyö, D.; Kahler, D.J.; Li, D.; Agmon, N.; Wudzinska, A.; Keegan, S.; Bader, J.S.; Yun, C.; et al. BRCA1 and S Phase DNA Repair Pathways Restrict LINE-1 Retrotransposition in Human Cells. Nat. Struct. Mol. Biol. 2020, 27, 179–191.
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-PiRNA Pathway Provides an Adaptive Defense in the Transposon Arms Race. Science 2007, 318, 761–764.
- Hancks, D.C.; Kazazian, H.H. Roles for Retrotransposon Insertions in Human Disease. Mob. DNA 2016, 7, 9.
- Kohlrausch, F.B.; Berteli, T.S.; Wang, F.; Navarro, P.A.; Keefe, D.L. Control of LINE-1 Expression Maintains Genome Integrity in Germline and Early Embryo Development. Reprod. Sci. 2021.
- Kim, S.; Günesdogan, U.; Zylicz, J.J.; Hackett, J.A.; Cougot, D.; Bao, S.; Lee, C.; Dietmann, S.; Allen, G.E.; Sengupta, R.; et al. PRMT5 Protects Genomic Integrity during Global DNA Demethylation in Primordial Germ Cells and Preimplantation Embryos. Mol. Cell 2014, 56, 564–579.
- Wu, Y.; Liu, W.; Chen, J.; Liu, S.; Wang, M.; Yang, L.; Chen, C.; Qi, M.; Xu, Y.; Qiao, Z.; et al. Nuclear Exosome Targeting Complex Core Factor Zcchc8 Regulates the Degradation of LINE1 RNA in Early Embryos and Embryonic Stem Cells. Cell Rep. 2019, 29, 2461–2472.e6.
- Lees-Murdock, D.J.; Walsh, C.P. DNA Methylation Reprogramming in the Germ Line. Adv. Exp. Med. Biol. 2008, 626, 1–15.
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A Unique Regulatory Phase of DNA Methylation in the Early Mammalian Embryo. Nature 2012, 484, 339–344.
- De Iaco, A.; Coudray, A.; Duc, J.; Trono, D. DPPA2 and DPPA4 Are Necessary to Establish a 2C-like State in Mouse Embryonic Stem Cells. EMBO Rep. 2019, 20, e47382.
- Kazazian, H.H.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370.
- Protasova, M.S.; Gusev, F.E.; Grigorenko, A.P.; Kuznetsova, I.L.; Rogaev, E.I.; Andreeva, T.V. Quantitative Analysis of L1-Retrotransposons in Alzheimer’s Disease and Aging. Biochemistry 2017, 82, 962–971.
- Kano, H.; Godoy, I.; Courtney, C.; Vetter, M.R.; Gerton, G.L.; Ostertag, E.M.; Kazazian, H.H. L1 Retrotransposition Occurs Mainly in Embryogenesis and Creates Somatic Mosaicism. Genes Dev. 2009, 23, 1303–1312.
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic Retrotransposition Alters the Genetic Landscape of the Human Brain. Nature 2011, 479, 534–537.
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 Retrotransposition in Human Neural Progenitor Cells. Nature 2009, 460, 1127–1131.
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A.; et al. Single-Neuron Sequencing Analysis of L1 Retrotransposition and Somatic Mutation in the Human Brain. Cell 2012, 151, 483–496.
- Evrony, G.D.; Lee, E.; Mehta, B.K.; Benjamini, Y.; Johnson, R.M.; Cai, X.; Yang, L.; Haseley, P.; Lehmann, H.S.; Park, P.J.; et al. Cell Lineage Analysis in Human Brain Using Endogenous Retroelements. Neuron 2015, 85, 49–59.
- Upton, K.R.; Gerhardt, D.J.; Jesuadian, J.S.; Richardson, S.R.; Sánchez-Luque, F.J.; Bodea, G.O.; Ewing, A.D.; Salvador-Palomeque, C.; van der Knaap, M.S.; Brennan, P.M.; et al. Ubiquitous L1 Mosaicism in Hippocampal Neurons. Cell 2015, 161, 228–239.
- Muotri, A.R.; Marchetto, M.C.N.; Coufal, N.G.; Oefner, R.; Yeo, G.; Nakashima, K.; Gage, F.H. L1 Retrotransposition in Neurons Is Modulated by MeCP2. Nature 2010, 468, 443–446.
- Jacob-Hirsch, J.; Eyal, E.; Knisbacher, B.A.; Roth, J.; Cesarkas, K.; Dor, C.; Farage-Barhom, S.; Kunik, V.; Simon, A.J.; Gal, M.; et al. Whole-Genome Sequencing Reveals Principles of Brain Retrotransposition in Neurodevelopmental Disorders. Cell Res. 2018, 28, 187–203.
- Bundo, M.; Toyoshima, M.; Okada, Y.; Akamatsu, W.; Ueda, J.; Nemoto-Miyauchi, T.; Sunaga, F.; Toritsuka, M.; Ikawa, D.; Kakita, A.; et al. Increased L1 Retrotransposition in the Neuronal Genome in Schizophrenia. Neuron 2014, 81, 306–313.
- Doyle, G.A.; Crist, R.C.; Karatas, E.T.; Hammond, M.J.; Ewing, A.D.; Ferraro, T.N.; Hahn, C.-G.; Berrettini, W.H. Analysis of LINE-1 Elements in DNA from Postmortem Brains of Individuals with Schizophrenia. Neuropsychopharmacology 2017, 42, 2602–2611.
- Liu, X.; Shimada, T.; Otowa, T.; Wu, Y.-Y.; Kawamura, Y.; Tochigi, M.; Iwata, Y.; Umekage, T.; Toyota, T.; Maekawa, M.; et al. Genome-Wide Association Study of Autism Spectrum Disorder in the East Asian Populations. Autism. Res. 2016, 9, 340–349.
- Lee, E.B.; Lee, V.M.-Y.; Trojanowski, J.Q. Gains or Losses: Molecular Mechanisms of TDP43-Mediated Neurodegeneration. Nat. Rev. Neurosci. 2011, 13, 38–50.
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 2006, 314, 130–133.
- Amador-Ortiz, C.; Lin, W.-L.; Ahmed, Z.; Personett, D.; Davies, P.; Duara, R.; Graff-Radford, N.R.; Hutton, M.L.; Dickson, D.W. TDP-43 Immunoreactivity in Hippocampal Sclerosis and Alzheimer’s Disease. Ann. Neurol. 2007, 61, 435–445.
- Nakashima-Yasuda, H.; Uryu, K.; Robinson, J.; Xie, S.X.; Hurtig, H.; Duda, J.E.; Arnold, S.E.; Siderowf, A.; Grossman, M.; Leverenz, J.B.; et al. Co-Morbidity of TDP-43 Proteinopathy in Lewy Body Related Diseases. Acta Neuropathol. 2007, 114, 221–229.
- Schwab, C.; Arai, T.; Hasegawa, M.; Yu, S.; McGeer, P.L. Colocalization of Transactivation-Responsive DNA-Binding Protein 43 and Huntingtin in Inclusions of Huntington Disease. J. Neuropathol. Exp. Neurol. 2008, 67, 1159–1165.
- Weihl, C.C.; Temiz, P.; Miller, S.E.; Watts, G.; Smith, C.; Forman, M.; Hanson, P.I.; Kimonis, V.; Pestronk, A. TDP-43 Accumulation in Inclusion Body Myopathy Muscle Suggests a Common Pathogenic Mechanism with Frontotemporal Dementia. J. Neurol. Neurosurg. Psychiatry 2008, 79, 1186–1189.
- Bose, J.K.; Huang, C.-C.; Shen, C.-K.J. Regulation of Autophagy by Neuropathological Protein TDP-43. J. Biol. Chem. 2011, 286, 44441–44448.
- Mitra, J.; Guerrero, E.N.; Hegde, P.M.; Liachko, N.F.; Wang, H.; Vasquez, V.; Gao, J.; Pandey, A.; Taylor, J.P.; Kraemer, B.C.; et al. Motor Neuron Disease-Associated Loss of Nuclear TDP-43 Is Linked to DNA Double-Strand Break Repair Defects. Proc. Natl. Acad. Sci. USA 2019, 116, 4696–4705.
- Liu, E.Y.; Russ, J.; Cali, C.P.; Phan, J.M.; Amlie-Wolf, A.; Lee, E.B. Loss of Nuclear TDP-43 Is Associated with Decondensation of LINE Retrotransposons. Cell Rep. 2019, 27, 1409–1421.e6.
- Morera, A.A.; Ahmed, N.S.; Schwartz, J.C. TDP-43 Regulates Transcription at Protein-Coding Genes and Alu Retrotransposons. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194434.
- Pereira, G.C.; Sanchez, L.; Schaughency, P.M.; Rubio-Roldán, A.; Choi, J.A.; Planet, E.; Batra, R.; Turelli, P.; Trono, D.; Ostrow, L.W.; et al. Properties of LINE-1 Proteins and Repeat Element Expression in the Context of Amyotrophic Lateral Sclerosis. Mob. DNA 2018, 9, 35.
- Hadlock, K.G.; Miller, R.G.; Jin, X.; Yu, S.; Reis, J.; Mass, J.; Gelinas, D.; Zhang, J.; McGrath, M.S. Elevated Rates of Antibody Reactivity to HML-2/HERV-K but Not Other Endogenous Retroviruses in ALS. Neurology 2004, 5, A37–A38.
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of Active Loci of a Human Endogenous Retrovirus in Neurons of Patients with Amyotrophic Lateral Sclerosis. Ann. Neurol. 2011, 69, 141–151.
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic Tau-Induced PiRNA Depletion Promotes Neuronal Death through Transposable Element Dysregulation in Neurodegenerative Tauopathies. Nat. Neurosci. 2018, 21, 1038–1048.
- Guo, C.; Jeong, H.-H.; Hsieh, Y.-C.; Klein, H.-U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880.
- Ittner, L.M.; Götz, J. Amyloid-β and Tau--a Toxic Pas de Deux in Alzheimer’s Disease. Nat. Rev. Neurosci. 2011, 12, 65–72.
- Rüb, U.; Stratmann, K.; Heinsen, H.; Seidel, K.; Bouzrou, M.; Korf, H.-W. Alzheimer’s Disease: Characterization of the Brain Sites of the Initial Tau Cytoskeletal Pathology Will Improve the Success of Novel Immunological Anti-Tau Treatment Approaches. J. Alzheimers Dis. 2017, 57, 683–696.
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A Protein Factor Essential for Microtubule Assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862.
- Nizynski, B.; Dzwolak, W.; Nieznanski, K. Amyloidogenesis of Tau Protein. Protein Sci. 2017, 26, 2126–2150.
- Ittner, A.; Ittner, L.M. Dendritic Tau in Alzheimer’s Disease. Neuron 2018, 99, 13–27.
- Hayflick, S.J.; Kurian, M.A.; Hogarth, P. Neurodegeneration with Brain Iron Accumulation. Handb. Clin. Neurol. 2018, 147, 293–305.
- Stanga, S.; Caretto, A.; Boido, M.; Vercelli, A. Mitochondrial Dysfunctions: A Red Thread across Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 3719.
- La Morgia, C.; Maresca, A.; Caporali, L.; Valentino, M.L.; Carelli, V. Mitochondrial Diseases in Adults. J. Intern Med. 2020, 287, 592–608.
- Missiroli, S.; Genovese, I.; Perrone, M.; Vezzani, B.; Vitto, V.A.M.; Giorgi, C. The Role of Mitochondria in Inflammation: From Cancer to Neurodegenerative Disorders. J. Clin. Med. 2020, 9, 740.
- Baeken, M.W.; Moosmann, B.; Hajieva, P. Retrotransposon Activation by Distressed Mitochondria in Neurons. Biochem. Biophys. Res. Commun. 2020, 525, 570–575.
- Giorgi, G.; Marcantonio, P.; Del Re, B. LINE-1 Retrotransposition in Human Neuroblastoma Cells Is Affected by Oxidative Stress. Cell Tissue Res. 2011, 346, 383–391.
- Whongsiri, P.; Pimratana, C.; Wijitsettakul, U.; Sanpavat, A.; Jindatip, D.; Hoffmann, M.J.; Goering, W.; Schulz, W.A.; Boonla, C. Oxidative Stress and LINE-1 Reactivation in Bladder Cancer Are Epigenetically Linked through Active Chromatin Formation. Free Radic Biol. Med. 2019, 134, 419–428.
- Yang, T.-C.; Wu, P.-C.; Chung, I.-F.; Jiang, J.-H.; Fann, M.-J.; Kao, L.-S. Cell Death Caused by the Synergistic Effects of Zinc and Dopamine Is Mediated by a Stress Sensor Gene Gadd45b—Implication in the Pathogenesis of Parkinson’s Disease. J. Neurochem. 2016, 139, 120–133.
- Ravel-Godreuil, C.; Massiani-Beaudoin, O.; Mailly, P.; Prochiantz, A.; Joshi, R.L.; Fuchs, J. Perturbed DNA Methylation by Gadd45b Induces Chromatin Disorganization, DNA Strand Breaks and Dopaminergic Neuron Death. iScience 2021, 24, 102756.
