Long noncoding RNAs (lncRNAs) are RNAs with a length of over 200 nucleotides that do not have protein-coding abilities. Recent studies suggest that lncRNAs are highly involved in physiological functions and diseases. lncRNAs HNF1α-AS1 and HNF4α-AS1 are transcripts of lncRNA genes HNF1α-AS1 and HNF4α-AS1, which are antisense lncRNA genes located in the neighborhood regions of the transcription factor (TF) genes HNF1α and HNF4α, respectively. HNF1α-AS1 and HNF4α-AS1 have been reported to be involved in several important functions in human physiological activities and diseases. In the liver, HNF1α-AS1 and HNF4α-AS1 regulate the expression and function of several drug-metabolizing cytochrome P450 (P450) enzymes, which also further impact P450-mediated drug metabolism and drug toxicity. In addition, HNF1α-AS1 and HNF4α-AS1 also play important roles in the tumorigenesis, progression, invasion, and treatment outcome of several cancers. Through interacting with different molecules, including miRNAs and proteins, HNF1α-AS1 and HNF4α-AS1 can regulate their target genes in several different mechanisms including miRNA sponge, decoy, or scaffold.
- lncRNA
- HNF1α-AS1
- HNF4α-AS1
- cancer
- cytochrome P450
1. Introduction
2. Neighborhood Antisense lncRNAs HNF1α-AS1 and HNF4α-AS1
2.1. Classification of lncRNAs Based on Genomic Locations
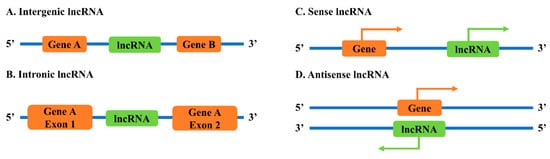
2.2. Neighborhood Antisense lncRNAs to Sense Genes
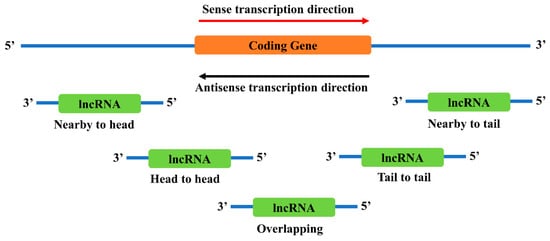
2.3. Neighborhood Antisense lncRNAs to TFs
2.4. Genomic Locations and Structures of HNF1α-AS1 and HNF4α-AS1
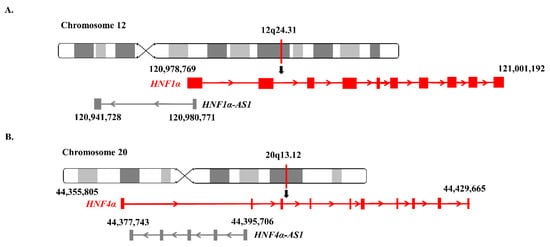
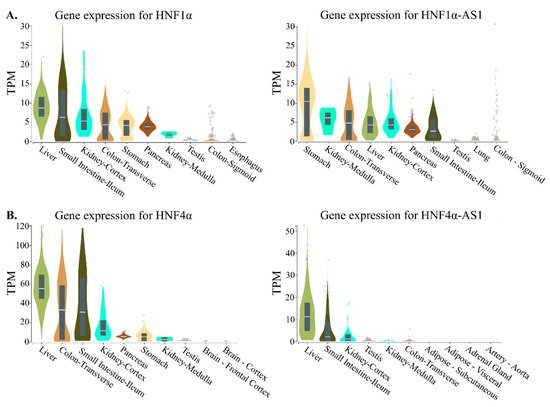
3. Relations of the Regulation of Expression between HNF1α and HNF1α-AS1 and between HNF4α and HNF4α-AS1
4. Functions of HNF1α-AS1 and HNF4α-AS1 in Human Physiology and Diseases
4.1. lncRNAs in Human Physiology and Diseases
4.2. Roles of HNF1α-AS1 and HNF4α-AS1 in Physiologic Functions, including Drug Metabolism
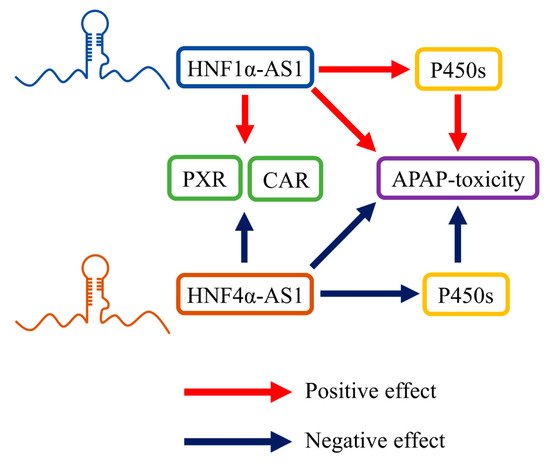
4.3. Role of HNF1α-AS1 and HNF4α-AS1 in the Progress of Human Diseases, including Cancers
References
- Szymanski, M.; Barciszewski, J. Beyond the proteome: Non-coding regulatory RNAs. Genome Biol. 2002, 3, 1–8.
- Choi, S.W.; Kim, H.W.; Nam, J.W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019, 20, 1853–1864.
- Chen, J.; Brunner, A.D.; Cogan, J.Z.; Nunez, J.K.; Fields, A.P.; Adamson, B.; Itzhak, D.N.; Li, J.Y.; Mann, M.; Leonetti, M.D.; et al. Pervasive functional translation of noncanonical human open reading frames. Science 2020, 367, 1140–1146.
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929.
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773.
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139.
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314.
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166.
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307.
- Guil, S.; Esteller, M. Cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol. 2012, 19, 1068–1075.
- Boque-Sastre, R.; Soler, M.; Oliveira-Mateos, C.; Portela, A.; Moutinho, C.; Sayols, S.; Villanueva, A.; Esteller, M.; Guil, S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 2015, 112, 5785–5790.
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol 2015, 89, 98–112.
- Lewandowski, J.P.; Lee, J.C.; Hwang, T.; Sunwoo, H.; Goldstein, J.M.; Groff, A.F.; Chang, N.P.; Mallard, W.; Williams, A.; Henao-Meija, J.; et al. The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat. Commun. 2019, 10, 5137.
- Li, W.; Shen, W.; Zhang, B.; Tian, K.; Li, Y.; Mu, L.; Luo, Z.; Zhong, X.; Wu, X.; Liu, Y.; et al. Long non-coding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b. Protein Cell 2020, 11, 161–186.
- Liu, B.; Ye, B.; Yang, L.; Zhu, X.; Huang, G.; Zhu, P.; Du, Y.; Wu, J.; Qin, X.; Chen, R. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat. Immunol. 2017, 18, 499.
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566.
- Numata, K.; Kiyosawa, H. Genome-wide impact of endogenous antisense transcripts in eukaryotes. Front. Biosci. 2012, 17, 300–315.
- Nishizawa, M.; Okumura, T.; Ikeya, Y.; Kimura, T. Regulation of inducible gene expression by natural antisense transcripts. Front. Biosci. 2012, 17, 938–958.
- Rosikiewicz, W.; Makalowska, I. Biological functions of natural antisense transcripts. Acta Biochim. Pol. 2016, 63, 665–673.
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural antisense transcripts: Molecular mechanisms and implications in breast cancers. Int. J. Mol. Sci. 2018, 19, 123.
- Wanowska, E.; Kubiak, M.R.; Rosikiewicz, W.; Makalowska, I.; Szczesniak, M.W. Natural antisense transcripts in diseases: From modes of action to targeted therapies. Wiley Interdiscip. Rev. RNA 2018, 9.
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343–346.
- Dempsey, J.; Zhang, A.; Cui, J.Y. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genom. 2018, 19, 834.
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227.
- Lin, S.; Zhang, L.; Luo, W.; Zhang, X. Characteristics of antisense transcript promoters and the regulation of their activity. Int. J. Mol. Sci. 2016, 17, 9.
- Uesaka, M.; Nishimura, O.; Go, Y.; Nakashima, K.; Agata, K.; Imamura, T. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genom. 2014, 15, 35.
- Herriges, M.J.; Swarr, D.T.; Morley, M.P.; Rathi, K.S.; Peng, T.; Stewart, K.M.; Morrisey, E.E. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014, 28, 1363–1379.
- Cao, Y.; Wang, J.; Lu, X.; Kong, X.; Bo, C.; Li, S.; Li, J.; Sun, X.; Wang, N.; Tian, K.; et al. Construction of a long noncoding RNAmediated transcription factor and gene regulatory triplet network reveals global patterns and biomarkers for ischemic stroke. Int. J. Mol. Med. 2020, 45, 333–342.
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117.
- Villegas, V.E.; Zaphiropoulos, P.G. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015, 16, 3251–3266.
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F., 3rd; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512.
- Usmanova, N.; Tomilin, N.; Zhivotovsky, B.; Kropotov, A. Transcription factor GABP/NRF-2 controlling biogenesis of mitochondria regulates basal expression of peroxiredoxin V but the mitochondrial function of peroxiredoxin V is dispensable in the dog. Biochimie 2011, 93, 306–313.
- Tani, H.; Torimura, M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem. Biophys. Res. Commun. 2013, 439, 547–551.
- Qi, W.; Li, Z.; Xia, L.; Dai, J.; Zhang, Q.; Wu, C.; Xu, S. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci. Rep. 2019, 9, 16185.
- Deng, Z.; Cao, P.; Wan, M.M.; Sui, G. Yin Yang 1: A multifaceted protein beyond a transcription factor. Transcription 2010, 1, 81–84.
- Wang, H.; Hertlein, E.; Bakkar, N.; Sun, H.; Acharyya, S.; Wang, J.; Carathers, M.; Davuluri, R.; Guttridge, D.C. NF-κB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 2007, 27, 4374–4387.
- Zhou, L.; Sun, K.; Zhao, Y.; Zhang, S.; Wang, X.; Li, Y.; Lu, L.; Chen, X.; Chen, F.; Bao, X.; et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015, 6, 10026.
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16.
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585.
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655.
- Ott, M.-O.; Rey-Campos, J.; Cereghini, S.; Yaniv, M. vHNF1 is expressed in epithelial cells of distinct embryonic origin during development and precedes HNF1 expression. Mech. Dev. 1991, 36, 47–58.
- Shih, D.Q.; Stoffel, M. Dissecting the transcriptional network of pancreatic islets during development and differentiation. Proc. Natl. Acad. Sci. USA 2001, 98, 14189–14191.
- Garrison, W.D.; Battle, M.A.; Yang, C.; Kaestner, K.H.; Sladek, F.M.; Duncan, S.A. Hepatocyte nuclear factor 4α is essential for embryonic development of the mouse colon. Gastroenterology 2006, 130, 19-e1.
- Chiang, J.Y. Hepatocyte nuclear factor 4α regulation of bile acid and drug metabolism. Expert Opin. Drug Metab. Toxicol. 2009, 5, 137–147.
- Shih, D.Q.; Bussen, M.; Sehayek, E.; Ananthanarayanan, M.; Shneider, B.L.; Suchy, F.J.; Shefer, S.; Bollileni, J.S.; Gonzalez, F.J.; Breslow, J.L. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 2001, 27, 375–382.
- Pontoglio, M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J. Am. Soc. Nephrol. 2000, 11, S140–S143.
- Pontoglio, M.; Sreenan, S.; Roe, M.; Pugh, W.; Ostrega, D.; Doyen, A.; Pick, A.J.; Baldwin, A.; Velho, G.; Froguel, P.; et al. Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J. Clin. Investig. 1998, 101, 2215–2222.
- Cheung, C.; Akiyama, T.E.; Kudo, G.; Gonzalez, F.J. Hepatic expression of cytochrome P450s in hepatocyte nuclear factor 1-alpha (HNF1alpha)-deficient mice. Biochem. Pharmacol. 2003, 66, 2011–2020.
- Kamiyama, Y.; Matsubara, T.; Yoshinari, K.; Nagata, K.; Kamimura, H.; Yamazoe, Y. Role of human hepatocyte nuclear factor 4α in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab. Pharm. 2007, 22, 287–298.
- Anik, A.; Catli, G.; Abaci, A.; Bober, E. Maturity-onset diabetes of the young (MODY): An update. J. Pediatric Endocrinol. Metab. 2015, 28, 251–263.
- Babeu, J.P.; Boudreau, F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J. Gastroenterol. 2014, 20, 22–30.
- Patitucci, C.; Couchy, G.; Bagattin, A.; Caneque, T.; de Reynies, A.; Scoazec, J.Y.; Rodriguez, R.; Pontoglio, M.; Zucman-Rossi, J.; Pende, M.; et al. Hepatocyte nuclear factor 1alpha suppresses steatosis-associated liver cancer by inhibiting PPARgamma transcription. J. Clin. Investig. 2017, 127, 1873–1888.
- Lu, Y.; Xu, D.; Peng, J.; Luo, Z.; Chen, C.; Chen, Y.; Chen, H.; Zheng, M.; Yin, P.; Wang, Z. HNF1A inhibition induces the resistance of pancreatic cancer cells to gemcitabine by targeting ABCB1. EBioMedicine 2019, 44, 403–418.
- Bonzo, J.A.; Patterson, A.D.; Krausz, K.W.; Gonzalez, F.J. Metabolomics identifies novel Hnf1α-dependent physiological pathways in vivo. Mol. Endocrinol. 2010, 24, 2343–2355.
- Takashima, Y.; Horisawa, K.; Udono, M.; Ohkawa, Y.; Suzuki, A. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci. 2018, 109, 3543–3553.
- Luo, Z.; Li, Y.; Wang, H.; Fleming, J.; Li, M.; Kang, Y.; Zhang, R.; Li, D. Hepatocyte nuclear factor 1A (HNF1A) as a possible tumor suppressor in pancreatic cancer. PLoS ONE 2015, 10, e0121082.
- Abel, E.V.; Goto, M.; Magnuson, B.; Abraham, S.; Ramanathan, N.; Hotaling, E.; Alaniz, A.A.; Kumar-Sinha, C.; Dziubinski, M.L.; Urs, S. HNF1A is a novel oncogene that regulates human pancreatic cancer stem cell properties. Elife 2018, 7, e33947.
- Ding, C.H.; Yin, C.; Chen, S.J.; Wen, L.Z.; Ding, K.; Lei, S.J.; Liu, J.P.; Wang, J.; Chen, K.X.; Jiang, H.L.; et al. The HNF1alpha-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer 2018, 17, 63.
- Chen, L.; Bao, Y.; Piekos, S.C.; Zhu, K.; Zhang, L.; Zhong, X.B. A Transcriptional Regulatory Network Containing Nuclear Receptors and Long Noncoding RNAs Controls Basal and Drug-Induced Expression of Cytochrome P450s in HepaRG Cells. Mol. Pharmacol. 2018, 94, 749–759.
- Wang, Y.; Yan, L.; Liu, J.; Chen, S.; Liu, G.; Nie, Y.; Wang, P.; Yang, W.; Chen, L.; Zhong, X. The HNF1α-regulated lncRNA HNF1α-AS1 is Involved in the Regulation of Cytochrome P450 Expression in Human Liver Tissues and Huh7 Cells. J. Pharmacol. Exp. Ther. 2019, 368, 353–362.
- Liu, H.T.; Liu, S.; Liu, L.; Ma, R.R.; Gao, P. EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Res. 2018, 78, 5877–5890.
- Guo, S.; Lu, H. Novel mechanisms of regulation of the expression and transcriptional activity of hepatocyte nuclear factor 4α. J. Cell. Biochem. 2019, 120, 519–532.
- Melissari, M.T.; Grote, P. Roles for long non-coding RNAs in physiology and disease. Pflug. Arch. Eur. J. Physiol. 2016, 468, 945–958.
- Kopp, F. Molecular functions and biological roles of long non-coding RNAs in human physiology and disease. J. Gene Med. 2019, 21, e3104.
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 2, e01749.
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726.
- Giroud, M.; Scheideler, M. Long Non-Coding RNAs in Metabolic Organs and Energy Homeostasis. Int. J. Mol. Sci. 2017, 18.
- Fan, Z.; Zhao, M.; Joshi, P.D.; Li, P.; Zhang, Y.; Guo, W.; Xu, Y.; Wang, H.; Zhao, Z.; Yan, J. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017, 45, 5720–5738.
- Sahlu, B.W.; Zhao, S.; Wang, X.; Umer, S.; Zou, H.; Huang, J.; Zhu, H. Long noncoding RNAs: New insights in modulating mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2020, 11, 16.
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208.
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261.
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; Laurent, G.S., III; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730.
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106.
- Chen, L.; Wang, P.; Manautou, J.E.; Zhong, X.B. Knockdown of Long Noncoding RNAs Hepatocyte Nuclear Factor 1alpha Antisense RNA 1 and Hepatocyte Nuclear Factor 4alpha Antisense RNA 1 Alters Susceptibility of Acetaminophen-Induced Cytotoxicity in HepaRG Cells. Mol. Pharmacol. 2020, 97, 278–286.
- Wang, Y.; Fang, Z.; Hong, M.; Yang, D.; Xie, W. Long-noncoding RNAs (lncRNAs) in drug metabolism and disposition, implications in cancer chemo-resistance. Acta Pharm. Sin. B 2020, 10, 105–112.
- Pan, J.J.; Xie, X.J.; Li, X.; Chen, W. Long Non-coding RNAs and Drug Resistance. Asian Pac. J. Cancer Prev. 2015, 16, 8067–8073.
- Ning, B.; Yu, D.; Yu, A.M. Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem. Pharm. 2019, 169, 113638.
- Yu, D.; Chen, J.; Chen, S.; Xu, L.; Wu, L.; Li, D.; Luo, J.; Jin, Y.; Zhao, Y.; Knox, B.; et al. Coordinated Regulation of UGT2B15 Expression by Long Noncoding RNA LINC00574 and hsa-miR-129-5p in HepaRG Cells. Drug Metab. Dispos. 2020, 48, 297–306.
- Li, D.; Wu, L.; Knox, B.; Chen, S.; Tolleson, W.H.; Liu, F.; Yu, D.; Guo, L.; Tong, W.; Ning, B. Long noncoding RNA LINC00844-mediated molecular network regulates expression of drug metabolizing enzymes and nuclear receptors in human liver cells. Arch. Toxicol. 2020.
- Yang, X.; Song, J.H.; Cheng, Y.; Wu, W.; Bhagat, T.; Yu, Y.; Abraham, J.M.; Ibrahim, S.; Ravich, W.; Roland, B.C.; et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 2014, 63, 881–890.
- Cai, L.; Lv, J.; Zhang, Y.; Li, J.; Wang, Y.; Yang, H. The lnc RNA HNF 1A-AS 1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J. Cell. Mol. Med. 2017, 21, 2654–2662.
- Dang, Y.; Lan, F.; Ouyang, X.; Wang, K.; Lin, Y.; Yu, Y.; Wang, L.; Wang, Y.; Huang, Q. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J. Surg. Oncol. 2015, 13, 302.
- Zhan, Y.; Li, Y.; Guan, B.; Wang, Z.; Peng, D.; Chen, Z.; He, A.; He, S.; Gong, Y.; Li, X. Long non-coding RNA HNF1A-AS1 promotes proliferation and suppresses apoptosis of bladder cancer cells through upregulating Bcl-2. Oncotarget 2017, 8, 76656.
- Fang, C.; Qiu, S.; Sun, F.; Li, W.; Wang, Z.; Yue, B.; Wu, X.; Yan, D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017, 410, 50–62.
- Muller, S.; Raulefs, S.; Bruns, P.; Afonso-Grunz, F.; Plotner, A.; Thermann, R.; Jager, C.; Schlitter, A.M.; Kong, B.; Regel, I.; et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol. Cancer 2015, 14, 94.
- Ma, Y.F.; Liang, T.; Li, C.R.; Li, Y.J.; Jin, S.; Liu, Y. Long non-coding RNA HNF1A-AS1 up-regulation in non-small cell lung cancer correlates to poor survival. Eur. Rev. Med. Pharm. Sci. 2016, 20, 4858–4863.
- Zhao, H.; Hou, W.; Tao, J.; Zhao, Y.; Wan, G.; Ma, C.; Xu, H. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/beta-catenin signaling pathway. Am. J. Transl. Res. 2016, 8, 3503–3512.
- Liu, Z.; Wei, X.; Zhang, A.; Li, C.; Bai, J.; Dong, J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem. Biophys. Res. Commun. 2016, 473, 1268–1275.
- Zhang, X.; Xiong, Y.; Tang, F.; Bian, Y.; Chen, Y.; Zhang, F. Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 96, 877–883.
- Wang, C.; Mou, L.; Chai, H.X.; Wang, F.; Yin, Y.Z.; Zhang, X.Y. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 89, 926–932.
- Wu, Y.; Liu, H.; Shi, X.; Yao, Y.; Yang, W.; Song, Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget 2015, 6, 9160–9172.
- Zhang, G.; An, X.; Zhao, H.; Zhang, Q.; Zhao, H. Long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion via regulating miR-17-5p in non-small cell lung cancer. Biomed. Pharm. 2018, 98, 594–599.
- Guo, X.; Zhang, Y.; Liu, L.; Yang, W.; Zhang, Q. HNF1A-AS1 Regulates Cell Migration, Invasion and Glycolysis via Modulating miR-124/MYO6 in Colorectal Cancer Cells. Oncotargets Ther. 2020, 13, 1507–1518.
- Feng, Z.; Wang, B. Long non-coding RNA HNF1A-AS1 promotes cell viability and migration in human bladder cancer. Oncol. Lett. 2018, 15, 4535–4540.
- Gottesman, M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002, 53, 615–627.
- Luo, X.; Wei, J.; Yang, F.-L.; Pang, X.-X.; Shi, F.; Wei, Y.-X.; Liao, B.-Y.; Wang, J.-L. Exosomal lncRNA HNF1A-AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA-34b/TUFT1 axis. Cancer Cell Int. 2019, 19, 323.
- Degli Esposti, D.; Hernandez-Vargas, H.; Voegele, C.; Fernandez-Jimenez, N.; Forey, N.; Bancel, B.; Le Calvez-Kelm, F.; McKay, J.; Merle, P.; Herceg, Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget 2016, 7, 31862.
- Haberman, Y.; BenShoshan, M.; Di Segni, A.; Dexheimer, P.J.; Braun, T.; Weiss, B.; Walters, T.D.; Baldassano, R.N.; Noe, J.D.; Markowitz, J.; et al. Long ncRNA Landscape in the Ileum of Treatment-Naive Early-Onset Crohn Disease. Inflamm. Bowel. Dis. 2018, 24, 346–360.
