Transfer RNAs (tRNAs) are essential adaptors that mediate translation of the genetic code. Modifications to tRNA are installed as post-transcriptional events at multiple locations on the tRNA structure by specialized tRNA-modifying enzymes and can occur at the 2’OH group of the ribose moiety as well as various positions of all A, C, G, and U bases. These modifications are diverse in their chemical structures and functional properties, and respond to nutritional and environmental factors.
- tRNA modifications
- translation
- tRNA
- thionucleoside
- translational capacity
- hypermodification
1. Abstract
tRNA undergoes a variety of post-transcriptional modifications. These chemical changes modulate the reactivity, structure, and stability of tRNA. Modifications to tRNA are important for interactions with aminoacyl tRNA synthetases (aaRSs), the ribosome, and messenger RNA sequences (mRNA), thereby affecting the proficiency of translational processes. The synthesis and reactivity of modified tRNAs is affected by environmental and nutritional factors, which lead to dynamic changes in the overall tRNA epitranscriptome. Consequently, tRNA modifications are involved in stress responses impacting translational efficiency and are used to elicit cellular changes at both the transcriptional and post-translational levels. Extracurricular functions imbued by tRNA modifications assign roles to these adaptor molecules as markers, sensor, and regulators of translational capacity.
2. Introduction
Transfer RNAs (tRNAs) are essential adaptors that mediate the decoding of genetic information. Functional tRNA molecules contain a myriad of modified nucleosides that are essential for structural stability and efficient protein synthesis [1]. To date, nearly 100 individual modifications have been reported in mature tRNA molecules [2]. These modifications are instated post-transcriptionally at multiple locations of the tRNA by specialized tRNA-modifying enzymes, and can occur at the 2’OH group of the ribose moiety and at various positions of all four RNA bases (A, G, C, U). These modifications are diverse in their chemical nature and functional properties, and range from the addition of a simple thio or methyl groups to the insertion of highly elaborate structures, such as in the case of queuosine (Q) [3][4][5][6]. Hypermodifications, multiple chemical alterations occurring within the same base, are also observed in tRNA as evidenced by the 5-methylaminomethyl-2-thiouridine (mnm
Transfer RNAs (tRNAs) are essential adaptors that mediate the decoding of genetic information. Functional tRNA molecules contain a myriad of modified nucleosides that are essential for structural stability and efficient protein synthesis [1]. To date, nearly 100 individual modifications have been reported in mature tRNA molecules [2]. These modifications are instated post-transcriptionally at multiple locations of the tRNA by specialized tRNA-modifying enzymes, and can occur at the 2’OH group of the ribose moiety and at various positions of all four RNA bases (A, G, C, U). These modifications are diverse in their chemical nature and functional properties, and range from the addition of a simple thio or methyl groups to the insertion of highly elaborate structures, such as in the case of queuosine (Q) [3–6]. Hypermodifications, multiple chemical alterations occurring within the same base, are also observed in tRNA as evidenced by the 5-methylaminomethyl-2-thiouridine (mnm
5
s
2
U) or the 2-methylthio-
N6
-isopentenyl adenosine (ms
2
i
6A) modifications [7]. The enzymes and the sequence of biosynthetic steps involved during the sequential or independent installation of modifications varies across phylogenetic groups and are subject to distinct mechanisms of regulation at the transcriptional and post-translational levels.
A) modifications [7]. The enzymes and the sequence of biosynthetic steps involved during the sequential or independent installation of modifications varies across phylogenetic groups and are subject to distinct mechanisms of regulation at the transcriptional and post-translational levels.
The canonical roles assigned to tRNA modifications have been generally categorized as important elements for translational accuracy and efficiency. The lack of certain modifications impairs the tRNA’s ability to interact with translation partners including aminoacyl tRNA synthetases (aaRSs), elongation factor-tu (Ef-Tu), the ribosome, and coding RNA sequences [8–10]. Modifications participating in interactions with translation components are present throughout the structure of tRNA (Figure 1). They serve as structural elements that promote the proper folding and conformation of these molecules, as well as functional elements that allow intramolecular chemical interactions within the tRNA and intermolecular interactions with translational partners. tRNA quality control checkpoints identify hypomodified tRNA and selectively target them for degradation, events of which modulate the tRNA pool and composition [11–13]. Additionally, the presence of certain modifications also serves as a primer for other tRNA-modifying enzymes during sequential biosynthetic schemes during the synthesis of modifications at other positions on the tRNA structure. Therefore, the degree and prevalence of these modifications impacts translational capacity through direct and indirect mechanisms.
The canonical roles assigned to tRNA modifications have been generally categorized as important elements for translational accuracy and efficiency. The lack of certain modifications impairs the tRNA’s ability to interact with translation partners including aminoacyl tRNA synthetases (aaRSs), elongation factor-tu (Ef-Tu), the ribosome, and coding RNA sequences [8][9][10]. Modifications participating in interactions with translation components are present throughout the structure of tRNA (Figure 1). They serve as structural elements that promote the proper folding and conformation of these molecules, as well as functional elements that allow intramolecular chemical interactions within the tRNA and intermolecular interactions with translational partners. tRNA quality control checkpoints identify hypomodified tRNA and selectively target them for degradation, events of which modulate the tRNA pool and composition [11][12][13]. Additionally, the presence of certain modifications also serves as a primer for other tRNA-modifying enzymes during sequential biosynthetic schemes during the synthesis of modifications at other positions on the tRNA structure. Therefore, the degree and prevalence of these modifications impacts translational capacity through direct and indirect mechanisms.
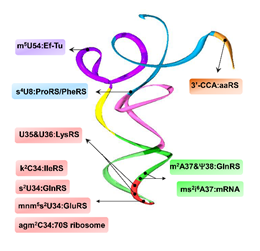
Figure 1. Modifications throughout the tRNA structure are important recognition elements for interaction with a variety of partners in translation. The tRNA structure (PDB 1u0b) shows domains color coded in accordance with Figure 1. m5U54 confers binding with T. aquaticus Ef-Tu [9]; s4U8 is necessary for structural stability impacting interactions with E. coli PheRS and ProRS [14]; U35/U36, along with mnm5s2U34, are major identity elements for E. coli LysRS binding [8]; agm2C34 interacts with the T. thermophilus 70S ribosome [15]; k2C34 is required for codon discrimination by IleRS in E. coli and B. subtilis [16]; s2U34, m2A37, and ψ38 are required for E. coli GlnRS substrate recognition and aminoacylation efficiency [17][18]; ms2i6A37 directly interacts with cognate mRNA bases in E, P and A sites of the ribosome [19]; all aaRSs require 3′-CCA moiety for tRNA charging [20].
Figure 1. Modifications throughout the tRNA structure are important recognition elements for interaction with a variety of partners in translation. The tRNA structure (PDB 1u0b) shows domains color coded in accordance with Figure 1. m5U54 confers binding with T. aquaticus Ef-Tu [9]; s4U8 is necessary for structural stability impacting interactions with E. coli PheRS and ProRS [14]; U35/U36, along with mnm5s2U34, are major identity elements for E. coli LysRS binding [8]; agm2C34 interacts with the T. thermophilus 70S ribosome [15]; k2C34 is required for codon discrimination by IleRS in E. coli and B. subtilis [16]; s2U34, m2A37, and ψ38 are required for E. coli GlnRS substrate recognition and aminoacylation efficiency [17,18]; ms2i6A37 directly interacts with cognate mRNA bases in E, P and A sites of the ribosome [19]; all aaRSs require 3′-CCA moiety for tRNA charging [20].
3. tRNA modifications respond to nutritional and environmental conditions
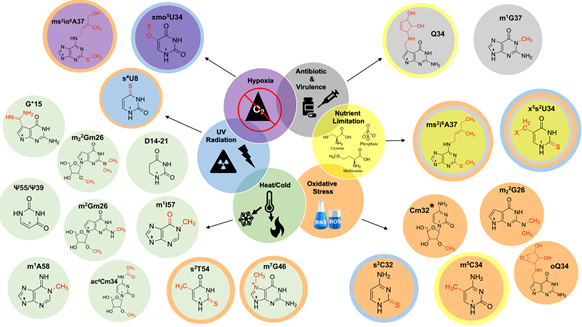
tRNA modifications are affected by a variety of nutritional and environmental stressors, and participate in a series of adaptive responses (Figure 2). These factors include nutrient limitation, changes in growth conditions such as extreme low/high growth temperatures, oxidative stress, respiratory requirements, local cellular pH fluctuations, and the presence of toxic compounds like antibiotics [21]. As a result, these stressors lead to dynamic changes in the tRNA epitranscriptome by either (1) directly participating in reactions with tRNA modifications or (2) by indirectly affecting the levels or activity of tRNA modifying enzymes. In both cases, these responses initiate the reprogramming of tRNA modifications and tRNA pool composition, leading to reduced growth rates and the upregulation of cellular stress response elements [22]. Thus, the occurrence of select post-transcriptional modifications on tRNA is proposed to contribute to adaptive mechanisms that enable the organism to survive during prolonged harsh growth conditions [23].
Figure 2. Schematic representation of tRNA modifications affected by nutritional and environmental stressors. Modifications affected by more than one condition are indicated with corresponding colored rings. *2-O’-methyladenosine (Am32) and 2-O’-methyluridine (Um32) are additionally affected by oxidative stress. The numbers “1” and “9” found within each base denote the β-glycosidic bond orientation found in the pyrimidine and purine bases, with their adjacent riboses.
4. Perspective on expanded functions of tRNA modifications
Extracurricular functions of tRNA modifications are credited to their critical roles during protein synthesis and how the degree and prevalence of these modified tRNAs impact translational competence and efficiency. Based on this premise, tRNA modifications affected by nutritional and environmental conditions are considered intermediates in cellular responses against a variety of stress factors. Whether serving as sensors, biomarkers, or regulatory elements, the reactivity of these modified molecules and their associated biosynthetic enzymes are central elements in promoting microbial fitness and adaptation across diverse habitats [24][25][26][24–26]. Understanding the mechanisms underlying the dynamic remodeling of the tRNA epitranscriptome in response to external stimuli provides an exciting avenue for identifying evolutionary determinants and microbial self-defense strategies that are distinct across phylogenetic groups. A comprehensive analysis of the non-canonical roles that tRNA modifications play in complex metabolic processes has recently been reviewed [27]. These emerging roles have clear implications in microbial physiology and metabolism. Moving forward, this field of research will continue to uncover the mechanisms by which modifications are able to respond to cellular stressors and potentially tune metabolic processes at the epigenetic and translational levels.
References:
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944, doi:10.1021/bi100408z.
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P. Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; Helm, M.; Bujnicki, J.M. MODOMICS: a database of RNA modification pathways. 2017 update. Nuc. Acids Res., 2018, 46, issue D1, D303.
- Black, K.A.; Santos, P.C.D. Abbreviated Pathway for Biosynthesis of 2-Thiouridine in Bacillus subtilis. Journal of Bacteriology 2015, 197, 1952–1962, doi:10.1128/JB.02625-14.
- Rajakovich, L.J.; Tomlinson, J.; Santos, P.C.D. Functional Analysis of Bacillus subtilis Genes Involved in the Biosynthesis of 4-Thiouridine in tRNA. Journal of Bacteriology 2012, 194, 4933–4940, doi:10.1128/JB.00842-12.
- Persson, B.C.; Gustafsson, C.; Berg, D.E.; Björk, G.R. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 3995–3998, doi:10.1073/pnas.89.9.3995.
- Miles, Z.D.; Myers, W.K.; Kincannon, W.M.; Britt, R.D.; Bandarian, V. Biochemical and Spectroscopic Studies of Epoxyqueuosine Reductase: A Novel Iron–Sulfur Cluster- and Cobalamin-Containing Protein Involved in the Biosynthesis of Queuosine. Biochemistry 2015, 54, 4927–4935, doi:10.1021/acs.biochem.5b00335.
- Zheng, C.; Black, K.A.; Dos Santos, P.C. Diverse Mechanisms of Sulfur Decoration in Bacterial tRNA and Their Cellular Functions. Biomolecules 2017, 7, 33, doi:10.3390/biom7010033.
- Cusack, S.; Yaremchuk, A.; Tukalo, M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996, 15, 6321–6334.
- Eargle, J.; Black, A.A.; Sethi, A.; Trabuco, L.G.; Luthey-Schulten, Z. Dynamics of Recognition between tRNA and Elongation Factor Tu. Journal of Molecular Biology 2008, 377, 1382–1405, doi:10.1016/j.jmb.2008.01.073.
- Jenner, L.B.; Demeshkina, N.; Yusupova, G.; Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010, 17, 555–560, doi:10.1038/nsmb.1790.
- Klaiman, D.; Amitsur, M.; Blanga-Kanfi, S.; Chai, M.; Davis, D.R.; Kaufmann, G. Parallel dimerization of a PrrC-anticodon nuclease region implicated in tRNALys recognition. Nucleic Acids Res 2007, 35, 4704–4714, doi:10.1093/nar/gkm494.
- Ogawa, T.; Inoue, S.; Yajima, S.; Hidaka, M.; Masaki, H. Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res 2006, 34, 6065–6073, doi:10.1093/nar/gkl629.
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA Decay Can Result from Lack of Nonessential Modifications. Molecular Cell 2006, 21, 87–96, doi:10.1016/j.molcel.2005.10.036.
- Thomas, G.; Thiam, K.; Favre, A. tRNA Thiolated Pyrimidines as Targets for Near-Ultraviolet-Induced Synthesis of Guanosine Tetraphosphate in Escherichia coli. European Journal of Biochemistry 1981, 119, 381–387, doi:10.1111/j.1432-1033.1981.tb05619.x.
- Voorhees, R.M.; Mandal, D.; Neubauer, C.; Köhrer, C.; RajBhandary, U.L.; Ramakrishnan, V. The structural basis for specific decoding of AUA by isoleucine tRNA on the ribosome. Nat. Struct. Mol. Biol. 2013, 20, 641–643, doi:10.1038/nsmb.2545.
- Muramatsu, T.; Nishikawa, K.; Nemoto, F.; Kuchino, Y.; Nishimura, S.; Miyazawa, T.; Yokoyama, S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 1988, 336, 179–181, doi:10.1038/336179a0.
- Rodriguez-Hernandez, A.; Spears, J.L.; Gaston, K.W.; Limbach, P.A.; Gamper, H.; Hou, Y.-M.; Kaiser, R.; Agris, P.F.; Perona, J.J. Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position. J Mol Biol 2013, 425, 3888–3906, doi:10.1016/j.jmb.2013.05.018.
- Enlisch-Peters, S.; Conley, J.; Plumbridge, J.; Leptak, C.; Söll, D.; Rogers, M.J. Mutant enzymes and tRNAs as probes of the glutaminyl-tRNA synthetase: tRNAGln interaction. Biochimie 1991, 73, 1501–1508, doi:10.1016/0300-9084(91)90184-3.
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate Translation of the Genetic Code Depends on tRNA Modified Nucleosides. J. Biol. Chem. 2002, 277, 16391–16395, doi:10.1074/jbc.M200253200.
- Sprinzl, M.; Cramer, F. The -C-C-A End of tRNA and Its Role in Protein Biosynthesis. In Progress in Nucleic Acid Research and Molecular Biology; Cohn, W.E., Ed.; Academic Press, 1979; Vol. 22, pp. 1–69.
- Shimizu, K. Regulation Systems of Bacteria such as Escherichia coli in Response to Nutrient Limitation and Environmental Stresses. Metabolites 2013, 4, 1–35, doi:10.3390/metabo4010001.
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nature Communications 2016, 7, 13302, doi:10.1038/ncomms13302.
- Alings, F.; Sarin, L.P.; Fufezan, C.; Drexler, H.C.A.; Leidel, S.A. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 2015, 21, 202–212, doi:10.1261/rna.048199.114.
- Ashraf, S.S.; Guenther, R.; Agris, P.F. Orientation of the tRNA anticodon in the ribosomal P-site: quantitative footprinting with U33-modified, anticodon stem and loop domains. RNA 1999, 5, 1191–1199.
- Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. Structure of a Ribonucleic Acid. Science 1965, 147, 1462–1465, doi:10.1126/science.147.3664.1462.
- Persson, B.C. Modification of tRNA as a regulatory device. Molecular Microbiology 1993, 8, 1011–1016, doi:10.1111/j.1365-2958.1993.tb01645.x.
- Edwards, A.M.; Addo, M.A.; Dos Santos, P.C. Extracurricular Functions of tRNA Modifications in Microorganisms. Genes 2020, 11, 907, doi:10.3390/genes11080907.