Breast cancer is the most frequent neoplasm in females. It is a heterogenous entity, classified into intrinsic subtypes based on gene expression data and in corresponding clinical subtypes based on the determination of hormone receptor expression and proliferative activity estimated from ki67 by immunohistochemistry. As for other tumors, the metabolism of breast tumors depends on aerobic glycolysis ("Warburg-effect") and the capability for effective biosynthesis of proteins. Quantity and quality of protein biosynthesis is mainly controlled in the initiation phase of translation, which is characterized by a complex interaction of eucaryotic initiation factors with the mRNA and ribosomal proteins to form a translationally active ribosome. Thus the eIF subunit composition varies from cancer to cancer and is a key factor for determining the cancer cell´s proteome. eIFs can therefore become a suitable anti-cancer drug target. We here summarize the current knowledge on eIF expression and prognostic impact in breast cancer.
- breast cancer
- eucaryotic intitation factors
- prognosis
Breast carcinoma (BC) remains one of the most serious health problems. It is a heterogeneous entity, and mainly classified according to receptor status for estrogen (ER), progesterone (PR) and egf (HER2/Neu), as well as the proliferation marker ki67. Gene expression in eukaryotes is regulated at the level of both gene transcription and translation, where eukaryotic initiation factors (eIFs) are key regulators of protein biosynthesis. Aberrant translation results in an altered cellular proteome, and this clearly effects cell growth supporting tumorigenesis. The relationship between various eIFs and BC entities, as well as the related regulatory mechanisms, has meanwhile become a focus of scientific interest. Here, we give an overview on the current research state of eIF function, focusing on BC.
1. Introduction
Worldwide, breast carcinoma (BC) remains one of the most serious health problems. The global annual incidence of reported BC cases is about 1,700,000. It is expected that incidence and mortality rates will increase significantly in the next 5–10 years [1]. Up to now, the median survival time in the advanced stage cohorts is still low (median survival time: 24 months) [2]. In developing countries, the BC rates are disproportionately high, and are estimated to increase to 55% in incidence, and to 58% in mortality, in the next 20 years [3]. BC is a heterogeneous entity, varying in hormone receptor status and expressions of human epidermal growth factor receptor (HER2) [4]. These markers, together with proliferation (as determined by ki67 expression or mitotic counts) and HER2 are used to assign BC to subtypes, and to predict the response to targeted therapies.
Additionally, BC can be classified according to gene expression analysis into four major molecular subtypes, namely luminal A, luminal B, HER2-enriched, and basal like [5,6][5][6]. From formalin-fixed paraffin-embedded (FFPE) BC samples, one can directly determine the four original subtypes, by applying a multigene assay i.e., the pam50 gene expression signature [7]. However, in clinical pathological diagnosis, we usually apply immunohistochemistry (IHC) to distinguish the four subtypes, by determining the status of the estrogen receptor (ER), progesterone receptor (PR), HER2 and Ki67 in BC tissue. The different expression status of these four parameters can used to be catalogued the subtypes. Luminal A-like subtype (approximate 40%) displays ER+ or PR+, or both, HER2− and low Ki67; luminal B-like subtype (approximate 40%) shows ER+ or PR+, or both, HER2− and high Ki67; HER2 subtype is divided into non-luminal (HER2+ and ER− and PR− (or luminal (HER2+ and ER+ or PR+, or both); basal-like subtype (HER2− and ER− and PR−, also called triple negative breast cancer). Up to now, at least ten different molecular subtypes have been identified using gene copy number and expression analyses [8]. These molecular subtypes are then used to guide systemic therapy for BC.
BC subtypes differ markedly in prognosis and in the therapeutic treatment. BC patients are usually treated by combination therapies of surgery, endocrine therapy, chemotherapy and radiotherapy. ER-positive BC patients are usually treated with five years of adjuvant endocrine therapy (Tamoxifen for premenopausal women or aromatase inhibitor or a combination of both after menopause). For preventing relapse in hormone-positive BC, patients might accept an extended endocrine therapy beyond five years [9]. Targeted therapy has substantially contributed to progress in BC therapy. Correspondingly, improved personalized treatments have minimized the side effects and improved overall survival rates. However, there is an annual increase in resistance rates to targeted drugs, depending on intra-tumor heterogeneity, adaptive processes and the patient’s genetic variation [3]. Even when targeted therapeutic strategies are applied, all breast cancer types might confront the oncologist with developing drug resistance. BC could resist multiple treatment strategies, such as chemotherapy, endocrine therapy or monoclonal antibody therapy. Hence, the current available targeted therapies are often limited.
Eukaryotic gene expression is mainly regulated at the level of transcription and translation. The deterioration of protein biosynthesis, divided into initiation, elongation, termination and ribosomal recycling, leads to an abnormal cellular proteome that can cause uncontrolled cell growth, as far as carcinogenesis is concerned [10]. Of the four stages of the translational cascade, initiation is the pivotal rate-limiting step. The eukaryotic initiation factors (eIFs) act as protein complexes called eIF1, eIF2, eIF3, eIF4, eIF5 and eIF6.
Indeed, previous research has shown that the dysregulation of several eIFs are related to carcinogenesis and cancer progression [11–15][11][12][13][14][15]. In this review, we will emphasize the contribution of eIFs to breast carcinogenesis, and provide a theoretical basis for new-targeted therapies.
2. Eukaryotic Translation Initiation Factors Dependencies in Breast Cancer
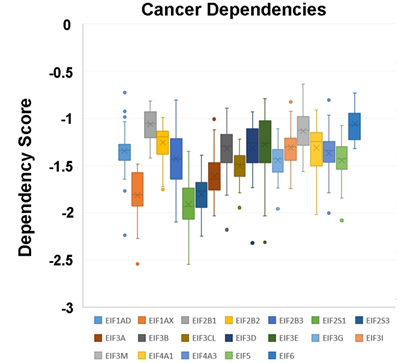
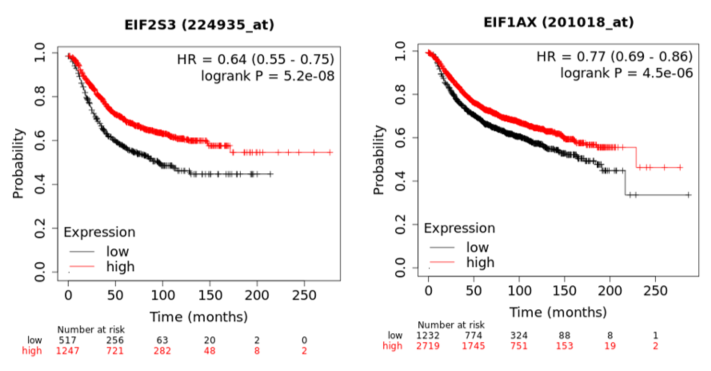
In a recent publication, a strategy to identify putative drug targets based on the genetic manipulation of cancer cell lines has been developed [85,86][16][17]. For this approach, cell lines have either been manipulated by systematic RNA-interference or CRIPR/CAS9 knockdown of genes. In order to assess the importance of eIFs for breast cancer cell lines, we have downloaded the data for the CRISPR/CAS9 knockdown for the eIFs available. A cancer dependency score of zero indicates that a gene is not essential for a cell–line and a score of −1 represents the median of all common essential genes. We therefore selected eIFs that have a mean dependency score in breast cancer cell lines smaller than −1 (Figure 1). The lowest score was found for eIF1AX, eif2S1 (eIF2α), eIF2S3 (eIF2γ) and eIF3a. The importance of eIF2α and eIF3A for breast cancer is clear from the literature. Interestingly for eIF1AX and eIF2γ, published data are scarce and deduced from the cancer dependency data—these genes should be investigated in more detail. Survival data from the KMplotter indicate, indeed, a strong prognostic effect for eIF2S3 and eIF1AX [87][18] (Figure 2).
Figure 1. Cancer dependencies for the eIFs as taken from the depmapportal (https://depmap.org/portal/). Only eIFs exhibiting mean dependencies lower than −1 are shown.
Figure 2. KM-plotter result for eIF2S3 and eIF2S3 for relapse-free survival in breast cancer.
3. Discussion
eIFs regulate the assembly of the functional ribosomal complex. There are six eIF-complexes participating in translation initiation. Disorders of eIF expression by over-expression, down-regulation or phosphorylation lead to carcinogenesis or tumor progression. Disturbance of the translation initiation leads to an abnormal cellular proteome, followed by uncontrolled cell growth. This highly controlled translation initiation process has attracted the attention of researchers, leading to an increased interest in targeting cancer by modifying translation initiation. Recent research on eIFs in BC has significantly expanded our knowledge on the broad spectrum of different stimuli of mammary epithelial cells (Table 1), having an impact on protein biosynthesis.
Table 1. eIF subunits and differential expression in breast cancer cell lines.
|
Protein |
Differential Expression |
Ductal Cancer |
Lobular Cancer |
Cell Lines |
References |
|
eIF2α |
↓ (eIF2α-P↑) |
√ |
|
BMECs MCF-7 SkBr3 |
|
|
eIF 3a |
↑ |
√ |
|
|
[35][21] |
|
eIF 3c |
↓ |
√ |
|
BT474 MDA-MB-231 |
[38][22] |
|
eIF3f |
↓ |
|
|
MCF-7 |
[37][23] |
|
eIF3h |
↑ |
|
|
MCF-7 MDA436 SK-Br-3 |
[39][24] |
|
eIF4a |
↑ |
|
|
MCF-7 |
[64][25] |
|
eIF 4b |
↑ |
|
|
MCF-7 |
[64][25] |
|
eIF 4e |
↑ |
√ |
|
MDA-MB-231 MDA-MB 468 MCF-7 HMECs |
|
|
eIF 4g |
↑ |
√ |
|
|
[10] |
|
eIF 5 |
↑ |
√ |
|
|
[75][33] |
Abbr.: HMECs: human mammary epithelial cells; BMECs: bovine mammary epithelial cells. ↓: decrease; ↑: increase;
Both eIF1 and eIF1a are required in translation initiation, by changing the ribosomal structure to facilitate mRNA-scanning, in order to allow start codon recognition. eIF1 can be affected by different types of genotoxic stress signaling.
The structure of eIF2 is more complex with regard to the heterotrimeric composition of the three subunits eIF2a, b and c. Increased phospho-eIF2α not only inhibits mRNA translation initiation, but also facilitates the translation of selected mRNAs. In chemotherapy, Trastuzumab in HER2+ BC can initiate a PKR/eIF2α-P pathway; eIF2α-P may be a potential biomarker for Trastuzumab efficacy. It is known that chemotherapy is associated with lots of side effects, limiting its use. The inhibition of eIF2α possibly suppresses BC growth and metastasis, which puts forward a new way of treating BC.
eIF3 participates in a few physiological and pathological processes. Some of the subunits emerge as new potential drug targets. eIF3a is also a potential oncogene involved in cancer occurrence, metastasis and therapy response. According to the physiological and pathological function, eIF3a represents a potential drug target. We need more efforts to improve the usability of currently available molecules or to design novel small molecule eIF3a regulators.
Among all the subunits of eIF4, eIF4E is of very high concern. eIF4E activity is regulated through the over-expression or phosphorylation of its binding protein 4E-BP1 in female BC. Phospho-4E-BP1 is a potential candidate for therapies directed towards its upstream kinase (mTOR). Moreover, the knockdown of eIF4E has also been shown to reduce BC cell migration and invasion.
As auxiliary proteins, participating in stimulating specific processes, the roles of eIF5, eIF5A and eIF5B in BC have not yet been well described during the protein translation cycle. Acting as a ribosomal anti-association factor, eIF6 has never been researched thoroughly either.
eIFs principally act as regulators of the translation initiation stage, a highly critical step, of utmost interest for targeting cancer. More detailed studies on eIFs are needed to provide further insights into the translation process in malignancies, which could provide important clues for the treatment of BC. Further studies on small molecule disruptors of eIFs or respective subunits will contribute to gaining a better understanding of the role of translational control, and will be one of the main focuses of research in the coming years.
References
- Oeffinger, K.C.; Fontham, E.T.; Etzioni, R.; Herzig, A.; Michaelson, J.S.; Shih, Y.C.; Walter, L.C.; Church, T.R.; Flowers, C.R.; LaMonte, S.J.; et al. Breast Cancer Screening for Women at Average Risk: 2015 Guideline Update from the American Cancer Society. JAMA 2015, 314, 1599–1614.
- Greaney, M.L.; Sprunck-Harrild, K.; Ruddy, K.J.; Ligibel, J.; Barry, W.T.; Baker, E.; Meyer, M.; Emmons, K.M.; Partridge, A.H. Study protocol for Young & Strong: A cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health 2015, 15, 37.
- Anastasiadi, Z.; Lianos, G.D.; Ignatiadou, E.; Harissis, H.V.; Mitsis, M. Breast cancer in young women: An overview. Updates Surg. 2017, 69, 313–317.
- Alqaisi, A.; Chen, L.; Romond, E.; Chambers, M.; Stevens, M.; Pasley, G.; Awasthi, M.; Massarweh, S. Impact of estrogen receptor (ER) and human epidermal growth factor receptor-2 (HER2) co-expression on breast cancer disease characteristics: Implications for tumor biology and research. Breast Cancer Res. Treat. 2014, 148, 437–444.
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752.
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874.
- Vieira, A.F.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests-Emergent Clinical Biomarkers. Front. Med. 2018, 5, 248.
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352.
- Glassman, D.; Hignett, S.; Rehman, S.; Linforth, R.; Salhab, M. Adjuvant Endocrine Therapy for Hormone-positive Breast Cancer, Focusing on Ovarian Suppression and Extended Treatment: An Update. Anticancer Res. 2017, 37, 5329–5341.
- Silvera, D.; Arju, R.; Darvishian, F.; Levine, P.H.; Zolfaghari, L.; Goldberg, J.; Hochman, T.; Formenti, S.C.; Schneider, R.J. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell Biol. 2009, 11, 903–908.
- Ganoza, M.C.; Kiel, M.C.; Aoki, H. Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 2002, 66, 460–485.
- Nasr, Z.; Robert, F.; Porco, J.A., Jr.; Muller, W.J.; Pelletier, J. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene 2013, 32, 861–871.
- Hamamura, K.; Minami, K.; Tanjung, N.; Wan, Q.; Koizumi, M.; Matsuura, N.; Na, S.; Yokota, H. Attenuation of malignant phenotypes of breast cancer cells through eIF2alpha-mediated downregulation of Rac1 signaling. Int. J. Oncol. 2014, 44, 1980–1988.
- Decarlo, L.; Mestel, C.; Barcellos-Hoff, M.H.; Schneider, R.J. Eukaryotic Translation Initiation Factor 4E Is a Feed-Forward Translational Coactivator of Transforming Growth Factor beta Early Protransforming Events in Breast Epithelial Cells. Mol. Cell. Biol. 2015, 35, 2597–2609.
- Kelly, N.J.; Varga, J.F.A.; Specker, E.J.; Romeo, C.M.; Coomber, B.L.; Uniacke, J. Hypoxia activates cadherin-22 synthesis via eIF4E2 to drive cancer cell migration, invasion and adhesion. Oncogene 2018, 37, 651–662.
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.
- Yu, C.; Mannan, A.M.; Yvone, G.M.; Ross, K.N.; Zhang, Y.L.; Marton, M.A.; Taylor, B.R.; Crenshaw, A.; Gould, J.Z.; Tamayo, P.; et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol. 2016, 34, 419–423.
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731.
- Darini, C.; Ghaddar, N.; Chabot, C.; Assaker, G.; Sabri, S.; Wang, S.; Krishnamoorthy, J.; Buchanan, M.; Aguilar-Mahecha, A.; Abdulkarim, B.; et al. An integrated stress response via PKR suppresses HER2+ cancers and improves trastuzumab therapy. Nat. Commun. 2019, 10, 2139.
- Maranto, C.; Perconti, G.; Contino, F.; Rubino, P.; Feo, S.; Giallongo, A. Cellular stress induces cap-independent alpha-enolase/MBP-1 translation. FEBS Lett. 2015, 589, 2110–2116.
- Marchetti, A.; Buttitta, F.; Pellegrini, S.; Bertacca, G.; Callahan, R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int. J. Oncol. 2001, 18, 175–179.
- Zhao, W.; Li, X.; Wang, J.; Wang, C.; Jia, Y.; Yuan, S.; Huang, Y.; Shi, Y.; Tong, Z. Decreasing Eukaryotic Initiation Factor 3C (EIF3C) Suppresses Proliferation and Stimulates Apoptosis in Breast Cancer Cell Lines Through Mammalian Target of Rapamycin (mTOR) Pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 4182–4191.
- Cuesta, R.; Berman, A.Y.; Alayev, A.; Holz, M.K. Estrogen receptor alpha promotes protein synthesis by fine-tuning the expression of the eukaryotic translation initiation factor 3 subunit f (eIF3f). J. Biol. Chem. 2019, 294, 2267–2278.
- Zhang, L.; Smit-McBride, Z.; Pan, X.; Rheinhardt, J.; Hershey, J.W. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J. Biol. Chem. 2008, 283, 24047–24060.
- Modelska, A.; Turro, E.; Russell, R.; Beaton, J.; Sbarrato, T.; Spriggs, K.; Miller, J.; Graf, S.; Provenzano, E.; Blows, F.; et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015, 6, e1603.
- Pettersson, F.; Yau, C.; Dobocan, M.C.; Culjkovic-Kraljacic, B.; Retrouvey, H.; Puckett, R.; Flores, L.M.; Krop, I.E.; Rousseau, C.; Cocolakis, E.; et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin. Cancer Res. 2011, 17, 2874–2884.
- Larsson, O.; Li, S.; Issaenko, O.A.; Avdulov, S.; Peterson, M.; Smith, K.; Bitterman, P.B.; Polunovsky, V.A. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 2007, 67, 6814–6824.
- Sonenberg, N. eIF4E, the mRNA cap-binding protein: From basic discovery to translational research. Biochem. Cell Biol. 2008, 86, 178–183.
- Robichaud, N.; del Rincon, S.V.; Huor, B.; Alain, T.; Petruccelli, L.A.; Hearnden, J.; Goncalves, C.; Grotegut, S.; Spruck, C.H.; Furic, L.; et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 2015, 34, 2032–2042.
- Li, Z.; Sun, Y.; Qu, M.; Wan, H.; Cai, F.; Zhang, P. Inhibiting the MNK-eIF4E-beta-catenin axis increases the responsiveness of aggressive breast cancer cells to chemotherapy. Oncotarget 2017, 8, 2906–2915.
- Zhou, X.; Tan, M.; Stone Hawthorne, V.; Klos, K.S.; Lan, K.H.; Yang, Y.; Yang, W.; Smith, T.L.; Shi, D.; Yu, D. Activation of the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancers. Clin. Cancer Res. 2004, 10, 6779–6788.
- Assouline, S.; Culjkovic, B.; Cocolakis, E.; Rousseau, C.; Beslu, N.; Amri, A.; Caplan, S.; Leber, B.; Roy, D.C.; Miller, W.H., Jr.; et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): A proof-of-principle clinical trial with ribavirin. Blood 2009, 114, 257–260.
- Jennings, M.D.; Pavitt, G.D. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 2010, 465, 378–381.