Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jason Zhu and Version 1 by wenwei lu.
By comparing phenotypes with genotypes based on genome-wide annotations, five tetracycline resistance genes, tet(M), tet(W/N/W), tet(L), tet(S), and tet(45), were detected in LAB. Multiple LAB strains without tetracycline resistance genes were found to be resistant to tetracycline at the currently recommended cutoff values.
- lactic acid bacteria
- tetracycline resistance
- minimum inhibitory concentration
- tetracycline resistance gene
- microbiological cutoff value
1. Introduction
Tetracyclines are widely used antibiotics in human medicine and animal husbandry owing to their broad-spectrum antibacterial activity, low production cost, and lack of serious adverse reactions [1]. However, with the extensive and unreasonable use of tetracyclines, bacterial tetracycline resistance has become a serious concern [2], and the acquisition of tetracycline resistance genes has been identified as the main cause of bacterial tetracycline resistance [3]. Most tetracycline resistance genes are dependent of the bacteria. So, usually the most frequent genes for Gram-negative bacteria are tet(A) and tet(B), which are relatively highly distributed [4,5][4][5]. Most tetracycline resistance genes are linked to transmissible plasmids, transposons, and conjugative transposons, which can quickly spread among bacteria in humans, animals, and the environment [6,7][6][7]. Accordingly, these genes pose a great threat to human and animal health [8].
Antimicrobial susceptibility testing is a traditional method of drug resistance testing. These testing methods include K-B disk diffusion, broth macrodilution, broth microdilution, agar dilution, and E-test [21,22][9][10]. These culture-based tests determine the growth of bacteria in the presence of antibiotics and evaluate bacterial resistance based on the drug resistance phenotype [23][11]. Microbiological cutoff values (MCOFFs) are usually used as the interpretation criteria to identify antibiotic resistance and to differentiate strains with acquired resistance from susceptible strains [24][12]. However, the cutoff values of some LAB species have not yet been determined. The EFSA guidelines classify these LAB species according to their fermentation type and determine the cutoff values based on their fermentation type [20][13].
The drug resistance of bacteria is usually determined by sequencing of coding genes. With the breakthrough of genome-wide sequencing technology, researchers have developed a sequence alignment method to identify antimicrobial resistance genes through sequence similarity [25][14]. By comparing the nucleic acid sequence or protein sequence of a strain with the sequence in the antimicrobial resistance database, the ARGs in the genome of LAB strains can be quickly identified and characterized to evaluate their drug resistance and risk of transfer [26][15]. Furthermore, the transformation of drug resistance evaluation from phenotype-based to genotype-based was promoted by such comparisons.
2. Results
2.1. Determination of the MICs and Identification of the Resistance Phenotype
2. Determination of the MICs and Identification of the Resistance Phenotype
To explore the tolerance level of different species of LAB to tetracycline, the MICs of tetracycline were tested using the broth microdilution method for 478 LAB strains. The MICs of tetracycline for the two quality control strains in this study were within the quality control range, and the MICs for all strains are presented in Table S1. Differences were observed in the intra- and interspecies levels of tetracycline susceptibility. Lactiplantibacillus plantarum and L. reuteri had a wide MIC range that covered 8 twofold dilutions compared to the remaining species. Furthermore, L. paracasei, L. rhamnosus and L. crispatus covered 7 twofold dilutions, while L. johnsonii and L. (para)gasseri covered only 5–6 twofold dilutions (Figure 1a–g). The MICs for L. paracasei and L. rhamnosus were mainly between 0.5 and 2 μg/mL, while those for L. reuteri, L. (para)gasseri, L. johnsonii, and L. crispatus were mainly between 2 and 8 μg/mL. In particular, compared to other species, L. plantarum showed higher MICs, ranging from 8 to 32 μg/mL. The MICs for L. johnsonii, L. crispatus, and L. reuteri showed an obvious bimodal distribution, suggesting that these species may contain acquired tetracycline resistance genes.
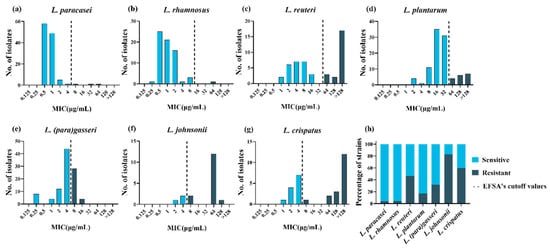

Figure 1. Minimum inhibitory concentration (MIC) distributions and drug resistance rate within 478 lactic acid bacteria strains. (a–g): Distribution of the MICs of tetracycline for eight lactic acid bacterial species. The black dotted lines represent epidemiological cutoff values reported by the European Food Safety Agency (EFSA). Sky blue represents phenotypically sensitive strains, and blue-black represents phenotypically resistant strains. (h) Distribution of tetracycline-resistant (black) and -susceptible (gray) strains according to the EFSA epidemiological cutoff values.
Phenotypic resistance was interpreted based on the MCOFFs reported by EFSA, which served as the interpretation criteria to distinguish susceptible strains free of phenotypically discoverable acquired resistance mechanisms from resistant strains. Generally, a strain was classified as susceptible when the MIC of a given antibiotic was not more than the established MCOFF. Herein, 23% (108/478) of the LAB strains were resistant to tetracycline; most L. johnsonii and L. crispatus strains showed tetracycline resistance, with resistance levels of 83% (15/18) and 60% (18/30), respectively. Lacticaseibacillus rhamnosus and L. paracasei showed the least common phenotypic resistance, with tetracycline resistance observed in 1% (1/68) and 3% (3/116) of the strains examined, respectively. Lactiplantibacillus plantarum, L. (para)gasseri, and L. reuteri exhibited tetracycline resistance levels of 17% (17/99), 32% (32/100), and 47% (22/47), respectively (Figure 1h).
2.2. Identification of ARGs and Their Correlation with Phenotype
3. Identification of ARGs and Their Correlation with Phenotype
The MCOFF is often used as an interpretation standard for the separation of sensitive strains from acquired resistance strains. If the MIC of one or more antibiotics for one strain is greater than the cutoff value, it is necessary to further explore its resistance mechanism at the genetic level [18][16]. Therefore, the genome sequences of 478 strains of LAB, which were tested for phenotypic resistance, were subjected to sequence alignment with CARD. Based on the selection standards, among the 478 strains, five tetracycline resistance-related genes encoded tetracycline target protection proteins (tet(W/N/W), tet(M), and tet(S)) and efflux pumps (tet(45) and tet(L)).
To identify the resistance determinants of tetracycline-resistant strains, wresaerchers analyzed the association between phenotypically resistant strains and their genotypes (Table 1). Among the three tetracycline-resistant strains of L. paracasei, the strain FCQHC12L3 was characterized by the presence of tet(M). However, no tetracycline resistance gene was found in the remaining two phenotypically resistant strains. Among the 17 tetracycline-resistant strains of L. plantarum, six were characterized by the presence of tet(M), while QHLJZD13-L6 was characterized by the presence of tet(S), and no related resistance genes were detected in 10 strains. Among the 22 tetracycline-resistant strains of L. reuteri, two strains were characterized by the presence of tet (M) and tet(L), whereas two strains were characterized by the presence of tet (W/N/W) and tet(L). Three strains and eleven strains harbored only tet(M) or tet(W/N/W), respectively. Furthermore, strain FYNLJ83L8 was characterized by the presence of tet(45), and three strains were not associated with resistance genes. Across the 15 tetracycline-resistant strains of L. johnsonii, the gene tet(W/N/W) was found in 11 resistant strains. In addition, FHNXY70M2 was characterized by the presence of tet(W/N/W) and tet (L), and the remaining three strains did not display resistance-related genes. Of the 18 resistant strains of L. crispatus, tet (M) was found in two resistant strains, and tet(W/N/W) was found in four resistant strains. Ten strains were characterized by the presence of tet(W/N/W) and tet(L), and the strain FHNXY70M14 was characterized by the presence of tet(M), tet(W/N/W), and tet(L). However, no resistance-related genes were detected in strain FHNXY56M7. No tetracycline resistance genes were found in any of the 32 tetracycline-resistant strains of L. (para)gasseri and one tetracycline-resistant strain of L. rhamnosus. In particular, two strains (L. plantarum RS41-7 and L. gasseri FHNFQ34_L1) harbored a tet(M) gene but were sensitive to tetracycline. Furthermore, the tet(M) sequence of the two strains had obvious deletions in the functional sites, resulting in the loss of resistance function (Figure 2). Therefore, in this study, we did not classify these two strains as carriers of the gene tet(M). In brief, through genotypic and phenotypic association analysis, these five tetracycline resistance genes could explain the resistance phenotypes of 33% (1/3) of L. paracasei, 41% (7/17) of L. plantarum, 80% (12/15) of L. johnsonii, 94% (17/18) of L. crispatus, and 86% (19/22) of L. reuteri strains. However, resistance phenotypes of 48% (52/108) of the resistant strains could not be explained based on their genotypes (Figure 3).

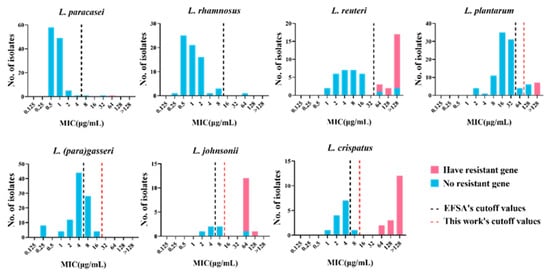

Figure 2. Multi-sequence alignment of tet(M) sequences of Lactiplantibacillus plantarum RS41-7 and Lactobacillus gasseri FHNFQ34_L1 (the bottom two sequences) with the tet(M) sequences of other strains in this study.

Figure 3. MIC distribution of 478 lactic acid bacteria strains with or without the tetracycline resistance gene. Blue: strains without resistance gene. Pink: strains with the tetracycline resistance gene. Black dotted line: cutoff value established by EFSA. Red dotted line: the new cutoff value established by this work.
Table 1. Association between tetracycline-resistant strains and tetracycline resistance genes in eight lactic acid bacterial species.
| Tetracycline Resistance Genes | Number of Tetracycline-Resistant Strains | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tet(M) | L. paracasei | (1), | L. plantarum | (6), | L. reuteri | (3), | L.crispatus | (2) | ||||||
| tet(W/N/W) | L. reuteri | (11), | L.johnsonii | (11), | L.crispatus | (4) | ||||||||
| tet(S) | L. plantarum | (1) | ||||||||||||
| tet(45) | L. reuteri | (1) | ||||||||||||
| tet(M) and tet(L) | L. reuteri | (2), | L.crispatus | (10) | ||||||||||
| tet(W/N/W) and tet(L) | L. reuteri | (2), | L.johnsonii | (1) | ||||||||||
| tet(M), tet(W/N/W), and tet(L) | L.crispatus | (1) | ||||||||||||
| No tetracycline resistance genes | L. paracasei | (2), | L. rhamnosus | (1), | L. plantarum | (10), | L. reuteri | (3), | L.johnsonii | (3), | L.crispatus | (1), | L. (para)gasseri | (32) |
2.3. Definition of New Susceptibility–Resistance Cutoff Values
3. Definition of New Susceptibility–Resistance Cutoff Values
The results of genotype-phenotype association analysis revealed that the genetic basis for the resistance of 48% (52/108) of the strains with tetracycline resistance phenotype could not be determined, including that of two strains of L. paracasei, one strain of L. rhamnosus, 10 strains of L. plantarum, one strain of L. crispatus, three strains of L. reuteri, three strains of L. johnsonii, and 32 strains of L. (para)gasseri. Lactobacillus (para)gasseri has a high drug resistance rate; however, the resistance determinants of all phenotypically resistant strains could not be identified. We speculate tThat the cutoff value based on the fermentation type may not be applicable to all species of LAB. Thus, it is recommended to develop MCOFFs at the species level. Lactobacillus (para)gasseri, L. crispatus, and L. johnsonii did not have species-specific cutoff values; therefore, we statistically analyzed the MIC frequency distribution of these species of Lactobacillus to establish species-specific TMCOFFs, which could better distinguish the resistant strains from the sensitive strains without acquired ARGs. Lacticaseibacillus paracasei, L. rhamnosus, L. plantarum, and L. reuteri had MCOFFs at the species level; however, 10% of L. plantarum had resistance phenotype but no resistance determinants. Therefore, wresearchers reformulated the cutoff value of L. plantarum to determine whether the strains containing resistance genes could be better distinguished from sensitive strains. Based on the MIC distribution of tetracycline of L. (para)gasseri, L. crispatus, L. johnsonii, and L. plantarum in this study, two different statistical approaches (Turnidge and Kronvall) and a “visual estimation” approach (eyeball method) were used to determine the new susceptibility–resistance cutoff values (Table 2).
Table 2. Comparison of tentative microbiological cutoff values (TMCOFFs) for tetracycline calculated using two statistical methods and the eyeball method.
| TMCOFFs Obtained Using the Indicated Method (%) | a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | EFSA Cut Off | Method of Turnidge et al. | b | Method of Kronvall |
Eyeball Method | Median for the Method |
|||
| L. (para)gasseri | 4 (68%) | 16 (100%) | 256 (100%) | 16 (100%) | 16 (100%) | ||||
| L. johnsonii | 4 (17%) | 32 (38%) | 16 (38%) | 16 (38%) | 16 (38%) | ||||
| L. crispatus | 4 (40%) | 8 (50%) | 16 (50%) | 16 (50%) | 16 (50%) | ||||
| L. plantarum | 32 (83%) | 64 (87%) | 64 (87%) | 64 (87%) | 64 (87%) | ||||
Based on the new cutoff values, all L. (para)gasseri strains were classified as sensitive, and the strains of L. crispatus containing resistance genes were distinguished from the sensitive strains without acquired ARGs. One strain belonging to L. johnsonii was not associated with the tetracycline resistance gene and was classified as phenotypically resistant. However, the MIC for this strain was found to be equivalent to that for another L. johnsonii strain containing the tetracycline resistance gene. Accordingly, we speculated that it contained potential tetracycline resistance genes. Therefore, the new cutoff value could completely distinguish the strains containing resistance genes from sensitive strains in L. johnsonii. The cutoff value of tetracycline for L. plantarum was newly formulated as 64 μg/mL, which is the same as that formulated by Flórez et al. [40][17]. However, the resistance determinants of six strains of L. plantarum still need to be further explored.
2.4. Prevalence and Distribution of Tetracycline Resistance Genes in LAB
To explore the distribution and prevalence of these five tetracycline resistance genes in LAB, wethis study performed statistical analysis on the detection of tetracycline resistance genes in eight species of LAB (Table 3). The most widely distributed tetracycline resistance gene in LAB was tet(M), which was detected in four LAB species, including L. paracasei, L. plantarum, L. reuteri, and L. crispatus. The genes tet(W/N/W) and tet(L) were detected in three species of LAB, including L. reuteri, L. johnsonii, and L. crispatus. The tet(S) gene was detected in only one strain of L. plantarum, and the tet (45) gene was found in only one strain of L. reuteri. The tet(W/N/W) gene was the most frequently detected tetracycline resistance gene in LAB; it was detected in 30 strains of LAB in this study. The genes tet(M), tet(L), tet(S), and tet(45) had detection frequencies of 26, 16, 1, and 1, respectively. Most types of the tetracycline resistance genes were detected in L. reuteri, including tet(M), tet(W/N/W), tet(L), and tet(45). Three of the tetracycline resistance genes were detected in L. crispatus, namely tet(M), tet(W/N/W), and tet(L). Two genes, tet(W/N/W) and tet(L), were detected in L. johnsonii. Two tetracycline resistance genes, tet(M) and tet(S), were found in L. plantarum. Among L. paracasei strains, only one harbored the tet(M) gene. No tetracycline resistance genes were found in any of the L. rhamnosus and L. (para)gasseri strains in this study. Herein, the tetracycline resistance gene was detected in 12% (56/478) of the strains. A total of 26 LAB strains contained only the tet(W/N/W) gene, while 12 LAB strains contained only the tet(M) gene. Interestingly, the gene tet(L) did not appear alone in LAB but was always detected together with other tetracycline resistance genes. The genes tet(M) and tet(L) were detected together in 12 strains of LAB. The genes tet(W/N/W) and tet(L) were detected concurrently in three strains of LAB. Three tetracycline resistance genes, tet(M), tet(W/N/W), and tet(L), were detected simultaneously in one L. crispatus strain. One strain of L. plantarum contained only the tet(S) gene, and one L. reuteri strain contained only the tet(45) gene.
Table 3. The detailed distribution of the detected tetracycline resistance genes in different lactic acid bacterial species.
| Species | Total Strain Number | TET | R | tet(M) | tet(W/N/W) | tet(L) | tet(S) | tet(45) |
|---|---|---|---|---|---|---|---|---|
| L. paracasei | 116 | 3 | 1 | 0 | 0 | 0 | 0 | |
| L. rhamnosus | 68 | 1 | 0 | 0 | 0 | 0 | 0 | |
| L. plantarum | 99 | 17 | 6 | 0 | 0 | 1 | 0 | |
| L. reuteri | 47 | 22 | 5 | 13 | 4 | 0 | 1 | |
| L. johnsonii | 18 | 15 | 0 | 12 | 1 | 0 | 0 | |
| L. crispatus | 30 | 18 | 13 | 5 | 11 | 0 | 0 | |
| L.(para)gasseri | 100 | 32 | 0 | 0 | 0 | 0 | 0 | |
| Total | 478 | 108 | 25 | 30 | 16 | 1 | 1 |
The genes of tet(M) and tet(W/N/W) are the most widely distributed tetracycline resistance genes in LAB. In order to further explore their phylogenetic relationship, multiple sequence alignments of a total of 25 tet(M) and 30 tet(W/N/W) gene sequences identified in this paper together with the same genotype sequences retrieved from the NCBI database were performed by ClustalW, and the phylogenetic tree was constructed using the Neighbor-Joining method by MEGA X (Figures S1 and S2). Most of the same species were in the same branch, suggesting that the genes had adaptive mutations when transferred to different species, and the tet(M) and tet(W/N/W) genes of LAB of the same species may have come from the same host.
3. Conclusions
Based on the MIC distribution data of 478 strains of LAB, LAB showed moderate resistance to tetracycline. Further, formulating the breakpoint value at the species level was found to be necessary. Therefore, the species-specific microbiological cutoff values for L. (para)gasseri, L. crispatus, and L. johnsonii against tetracycline were formulated, and new susceptibility-resistance cutoff values for L. plantarum were defined. The genes tet(M), tet(W/N/W), tet(L), tet(S), and tet(45) were the key resistance genes for the tetracycline resistance phenotype and were found to widely exist in LAB. The determination of antibiotic resistance in probiotic strains is related to food safety issues. The findings of this study provide certain guiding significance and reference values at the phenotype and genotype levels for the safe application of LAB in the food industry and the formulation of probiotic resistance evaluation standards.
References
- Jamal, M.; Shareef, M.; Sajid, S. Lincomycin and tetracycline resistance in poultry. Review. Matrix Sci. Pharma. 2017, 1, 33–38.
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrob. Agents Chemothe. 2018, 62, e00119-18.
- Tao, R.; Ying, G.-G.; Su, H.-C.; Zhou, H.-W.; Sidhu, J.P. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ. Pollut. 2010, 158, 2101–2109.
- Guarddon, M.; Miranda, J.M.; Rodríguez, J.A.; Vázquez, B.I.; Cepeda, A.; Franco, C.M. Real-time polymerase chain reaction for the quantitative detection of tetA and tetB bacterial tetracycline resistance genes in food. Int. J. Food Microbiol. 2011, 146, 284–289.
- Arredondo, A.; Àlvarez, G.; Nart, J.; Mor, C.; Blanc, V.; León, R. Detection and expression analysis of tet(B) in Streptococcus oralis. J. Oral Microbiol. 2019, 11, 1643204.
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992, 5, 387–399.
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456.
- Phillips, I.; Casewell, M.; Cox, T.; De Groot, B.; Friis, C.; Jones, R.; Nightingale, C.; Preston, R.; Waddell, J. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 2004, 53, 28–52.
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79.
- Kushiro, A.; Chervaux, C.; Cools-Portier, S.; Perony, A.; Legrain-Raspaud, S.; Obis, D.; Onoue, M.; van de Moer, A. Antimicrobial susceptibility testing of lactic acid bacteria and bifidobacteria by broth microdilution method and Etest. Int. J. Food Microbiol. 2009, 132, 54–58.
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based methods for detection of antibiotic resistance in agroecosystems: Advantages, challenges, and gaps in knowledge. J. Environ. Qual. 2016, 45, 432–440.
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J. Clin. Microbiol. 2019, 57, e01129-19.
- Additives, E.P.o.; Feed, P.o.S.u.i.A. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740.
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370.
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405-18.
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastro. Hepat. 2014, 11, 506–514.
- Flórez, A.B.; Egervärn, M.; Danielsen, M.; Tosi, L.; Morelli, L.; Lindgren, S.; Mayo, B. Susceptibility of Lactobacillus plantarum strains to six antibiotics and definition of new susceptibility–resistance cutoff values. Microb. Drug Resist. 2006, 12, 252–256.
More
