A novel mycovirus named Fusarium oxysporum alternavirus 1(FoAV1) was identified as infecting Fusarium oxysporum strain BH19, which was isolated from a fusarium wilt diseased stem of Lilium brownii. The genome of FoAV1 contains four double-stranded RNA (dsRNA) segments (dsRNA1, dsRNA 2, dsRNA 3 and dsRNA 4, with lengths of 3.3, 2.6, 2.3 and 1.8 kbp, respectively). Additionally, dsRNA1 encodes RNA-dependent RNA polymerase (RdRp), and dsRNA2- dsRNA3- and dsRNA4-encoded hypothetical proteins (ORF2, ORF3 and ORF4), respectively. A homology BLAST search, along with multiple alignments based on RdRp, ORF2 and ORF3 sequences, identified FoAV1 as a novel member of the proposed family "Alternaviridae". Evolutionary relation analyses indicated that FoAV1 may be related to alternaviruses, thus dividing the family "Alternavirida" members into four clades. In addition, we determined that dsRNA4 was dispensable for replication and may be a satellite-like RNA of FoAV1—and could perhaps play a role in the evolution of alternaviruses. Our results provided evidence for potential genera establishment within the proposed family "Alternaviridae". Additionally, FoAV1 exhibited biological control of Fusarium wilt.
A novel mycovirus named Fusarium oxysporum alternavirus 1(FoAV1) was identified as
infecting Fusarium oxysporum strain BH19, which was isolated from a fusarium wilt diseased stem
of Lilium brownii. The genome of FoAV1 contains four double-stranded RNA (dsRNA) segments
(dsRNA1, dsRNA 2, dsRNA 3 and dsRNA 4, with lengths of 3.3, 2.6, 2.3 and 1.8 kbp, respectively).
Additionally, dsRNA1 encodes RNA-dependent RNA polymerase (RdRp), and dsRNA2- dsRNA3-
and dsRNA4-encoded hypothetical proteins (ORF2, ORF3 and ORF4), respectively. A homology
BLAST search, along with multiple alignments based on RdRp, ORF2 and ORF3 sequences, identified
FoAV1 as a novel member of the proposed family “Alternaviridae”. Evolutionary relation analyses
indicated that FoAV1 may be related to alternaviruses, thus dividing the family “Alternaviridae”
members into four clades. In addition, we determined that dsRNA4 was dispensable for replication
and may be a satellite-like RNA of FoAV1—and could perhaps play a role in the evolution of
alternaviruses. Our results provided evidence for potential genera establishment within the proposed
family “Alternaviridae”. Additionally, FoAV1 exhibited biological control of Fusarium wilt. Our results
also laid the foundations for the further study of mycoviruses within the family “Alternaviridae”, and
provide a potential agent for the biocontrol of diseases caused by F. oxysporum.
1. Introduction
Mycoviruses are a group of viruses that infect filamentous fungi, yeasts and oomycetes. Until recently, the National Center of Biotechnology Information (NCBI) database contained more than 300 mycoviral sequences, divided into at least 19 families by the International Committee for Taxonomy of Viruses (ICTV)
[1]. Mycoviruses contain double-stranded RNA (dsRNA), positive single-stranded RNA (+ssRNA), negative single-stranded RNA (-ssRNA) and single-stranded DNA (ssDNA) as their genome
[2][3][4][2,3,4]. Double-stranded RNA (dsRNA) mycoviruses are classified into seven families:
Totiviridae, Endornaviridae, Partitiviridae, Chrysoviridae, Megabirnaviridae, Quadriviridae and
Reoviridae, and one genus,
Botybirnavirus [5][6][5,6].
Many mycoviruses affect fungal virulence and some can cause alterations in host phenotypes, reducing or enhancing virulence
[2][7][8][9][10][11][12][2,7,8,9,10,11,12]. This property of mycoviruses, which reduces the ability of their fungal hosts to cause disease, is termed hypovirulence
[2][4][2,4]. Hypovirulent mycoviruses have the potential to elicit biological control of plant pathogenic fungal diseases
[2][4][12][2,4,12]. Typical examples of hypovirulent mycovirus application include Cryphonectria hypovirus 1 (CHV1), which is used to control chestnut blight caused by
Cryphonectria parasitica in Europe, and the Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1), which is used to control stalk break caused by
Sclerotiorum sclerotiorum [13][14][15][16][13,14,15,16]. In recent years, many hypovirulence-associated mycoviruses have been found in other plant pathogenic fungi, such as
Botryosphaeria cinerea debilitation-related virus (BcDRV), Botryosphaeria dothidea Chrysovirus 1 (BdCV1), Rhizoctonia solani partitivirus 2 (RSPV2), Rhizoctonia solani endornavirus 1 (RsEV1) and Colletotrichum liriopes partitivirus 1 (ClPV1)
[17][18][19][20][17,18,19,20].
Further studies have revealed many novel viruses that differ obviously from already-known viruses. For example, Hammond et al. (2008) reported a novel mycovirus,
Aspergillus mycovirus 341 (AsV341), from
Aspergillus niger [21]. The closest known relative of the AsV341 was a totivirus, Sphaeropsis sapinea RNA virus 2 (SsRV2), from
Sphaeropsis sapinea [21]. SsRV2 contains only one genomic RNA segment (5202 bp) and is a typical totivirus, based on the sequence characteristics identified and phylogenetic analysis with other known totiviruses
[22]. Moreover, AsV341 contains four segments (between 1.5 and 3.6 kbp), and the amino acid identity and similarity levels between AsV341 and SsRV2 are only 15.2% and 27.8%, respectively
[21]. Thus, AsV341 is obviously different from the known totivirus, even though its closest relative is SsRV2
[21]. Aoki et al. (2009) reported a novel mycovirus, Alternaria alternata virus 1 (AaV1), from
Alternaria alternata, which is similar to AsV341, following an analysis of its sequence characteristics and multiple alignment. However, phylogenetic analysis showed that AaV1 and AsV341 did not cluster with any mycovirus family
[23]. In 2013, Kozlakidis et al.
[24] reported a novel mycovirus (Aspergillus foetidus virus (AfV)), which was similar to both AaV1 and AsV341, and proposed a new genus, Alternavirus, and a new family, “
Alternaviridae”, to accommodate the three viruses. Subsequently, several mycoviruses were sequenced and identified as potential members of the proposed “
Alternaviridae” family, including Fusarium poae alternavirus 1 (FpAV 1), Fusariumm graminerarum alternairus 1 (FgAV 1), Aspergillus heteromorphus alternavirus 1 (AheAV1) and Fusarium incarnatum alternavirus 1 (FiAV 1)
[25][26][27][28][25,26,27,28]. Until recently, mycovirus classification into the proposed family “
Alternaviridae” was based on phylogenetic analysis of RNA-dependent RNA polymerase (RdRp) sequences. Some genomes of viruses belonging to the proposed family “
Alternaviridae” contained three dsRNA segments, while others comprised four dsRNA segments.
Fusarium oxysporum is an important plant pathogenic fungus that causes vascular wilt in a wide variety of agricultural crop species
[29]. Biocontrol agents, such as plant growth- promoting rhizobacteria (PGPR) strains and nonpathogenic
F. oxysporum strains, have proven to be effective tools for controlling plant diseases caused by
F. oxysporum [30][31][30,31]. As more hypovirulence-associated mycoviruses have been investigated in attempts to control crop diseases, new insights and explorations regarding biological control using hypovirulence-associated mycoviruses to control plant diseases caused by
F. oxysporum [4][32][33][4,32,33] have become available. To our knowledge, six mycoviruses have been reported in
F. oxysporum. These include Fusarium oxysporum chrysovirus 1 (FoCV1), Fusarium oxysporum f.sp. dianthi mycovirus 1 (FodV1), classified as family
Chrysoviridae; Fusarium oxysporum f. sp. dianthi mitovirus 1 (FodMV1), classified as family
Narnaviridae; Fusarium oxysporum f. sp. dianthi hypovirus 2 (FodHV2), classified as family
Hypoviridae; Hadaka Virus 1 (HadV1), classified as family
Polymycoviridae; and Fusarium oxysporum ourmia-like virus 1 (FoOuLV1), classified as family
Botourmiaviridae [34][35][36][37][38][39][34,35,36,37,38,39]. Only FodV1 and FoOuLV1 are known to cause hypovirulence
[39][40][39,40].
2. Strains and Culture Conditions
Seven isolates of
F. oxysporium (BH19, BH3, BH19-4V, BH19-3V, MC-1, MC-1-4V and MC-1-3V) were used in the study. Strains BH19 and BH3 were isolated from a fusarium wilt-diseased stem of
Lilium brownii in Liaoning province of China in 2019. Strains BH19-4V and BH19-3V were single-ascospore isolates of strain BH19. MC-1 was a
F. oxysporum f. sp.
momordicae strain incorporating a hygromycin-resistance gene (hygromycin B phosphotransferase), which has a normal colony morphology and high virulence in its hosts. Strains MC-1-4V and MC-1-3V were derivative strains isolated from MC-1 following confrontation training with BH19-4V and BH19-3V. All strains were stored at −80 °C in glycerol and cultured on potato dextrose agar (PDA) medium at 28 °C. For dsRNA extraction, mycelium was cultured on a PDA plate covered with cellophane membranes at 28 °C for 4–5 days.
3. DsRNA Extraction and Purification
The extraction of dsRNA was performed as described previously
[39]. Strains were grown for 4–5 days on cellophane membranes on PDA medium. Fresh mycelia (1–2 g) were harvested to isolate dsRNA by selective absorption using cellulose powder CF-11 (Sigma–Aldrich, St. Louis, MO, USA), with nucleic acid co-precipitator added to improve the yield of dsRNA. The dsRNA was treated with RNase-free DNase I and S1 nuclease (Takara, Dalian, China). The dsRNAs were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and visualized with a gel documentation and image analysis system (InGenius LhR, Syngene, UK).
4. DsRNA Segments in F. oxysporum Strain BH19
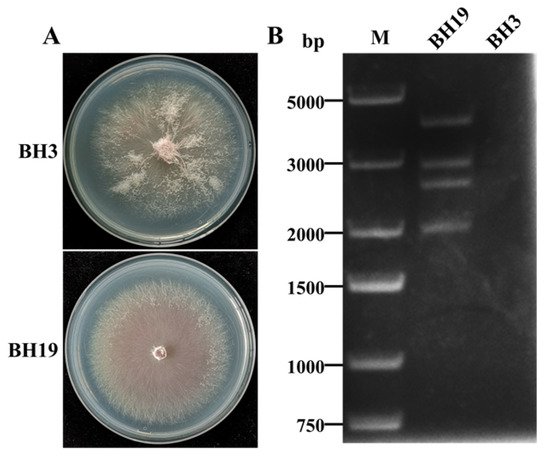
The
F. oxysporum strains BH19 and BH3 were both isolated from a fusarium wilt-diseased stem of
L. brownii in Liaoning Province, China. Compared to BH3, the colony morphology of BH19 was clearly defined with a small aerial mycelium (
Figure 1A). Strain BH19 contained four dsRNA segments (named dsRNA1, dsRNA2, dsRNA3 and dsRNA4) while no dsRNAs were found in strain BH3 (
Figure 1B). The four dsRNA bands had estimated sizes of 3.5 kbp (dsRNA1), 2.5 kbp (dsRNA2), 2.3 kbp (dsRNA3) and 1.8 kbp (dsRNA4), respectively. The segments were confirmed to be dsRNA in nature based on resistance to digestion with DNase I and S1 nuclease.
Figure 1. (A) Colony morphology of F. oxysporum strains BH3 and BH19. (B) Agarose gel electrophoresis of the dsRNA-enriched extract obtained by cellulose column chromatography from strains BH3 and BH19.
5. Molecular Characterization of FoAV1 in Strain BH19
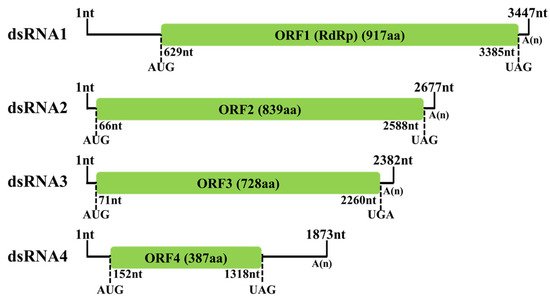
Following cDNA cloning and sequencing, the complete sequences of the four dsRNA segments found in strain BH19 were obtained, comprising a virus nominated as Fusarium oxysporum alternavirus 1 (FoAV1). The complete sequence of FoAV1 dsRNA1 (GenBank Accession No. MT659125) was 3447 nucleotides (nt) long with a GC content of 56.6%, and it contained one ORF (ORF1) that initiated at position nt 629 and terminated at position nt 3385. Based on the universal genetic code, this sequence potentially encoded 917 amino acid (aa) residues with a calculated molecular mass (
Mr) of 103.4 kDa. FoAV1 dsRNA2 (GenBank Accession No. MT659126) was 2677 nt in length with a GC content of 56.8%. It contained one ORF (ORF2) that initiated at position nt 66 and terminated at nt 2588, encoding 839 aa residues with an
Mr of 92.8 kDa. FoAV1 dsRNA3 (GenBank Accession No. MT659127) was 2382 nt in length with a GC content of 53.9%. It contained one ORF (ORF3) that initiated at position nt 71 and terminated at nt 2260, encoding 728 aa residues with an
Mr of 78.5 kDa. FoAV1 dsRNA4 (GenBank Accession No. MT659128) was 1873 nt in length with a GC content of 55.7%. It contained one ORF (ORF4) that initiated at position nt 152 and terminated at nt 1318, encoding 387 aa residues with an
Mr of 42.6 kDa. Additionally, all the four dsRNA segments contained poly (A) tails (
Figure 2).
Figure 2. Schematic representation of the FoAV1 genome. The open reading frames (ORFs) and coding loci are indicated by a green box and single dotted lines, respectively.