Sirtuins are key players for maintaining cellular homeostasis and are often deregulated in different human diseases. SIRT7 is the only member of mammalian sirtuins that principally resides in the nucleolus, a nuclear compartment involved in ribosomal biogenesis, senescence, and cellular stress responses. The ablation of SIRT7 induces global genomic instability, premature ageing, metabolic dysfunctions, and reduced stress tolerance, highlighting its critical role in counteracting ageing-associated processes.
- sirtuins
- deacetylation
- stress responses
- nucleolus
1. Mammalian Sirtuins: General Functions and Activation in Response to Stress
Sirtuins are highly conserved enzymes that principally act as NAD + -dependent histone/protein deacetylases although some members also possess mono-ADP ribosylation and other less characterized activities [1,2,3,4,5,6][1][2][3][4][5][6]. For deacetylation, sirtuins catalyze the transfer of the acetyl group from the substrate to NAD + , concomitant with the release of 2’–O-acetyl-ADP-ribose and nicotinamide (NAM). During mono-ADP ribosylation, the ADP-ribose (ADPR) is transferred from NAD + to the target protein and NAM is released ( Figure 1 a). Interestingly, NAM is a potent inhibitor of sirtuins, suggesting the presence of a finely controlled negative auto-regulatory loop, modulating the enzymatic activity ( Figure 1 a). In mammals, seven sirtuin members ( SIRT1 – SIRT7 ) have been described. These molecules share a conserved catalytic domain but differ substantially in their N-terminal and C-terminal sequences, which are fundamental for interactions with specific targets as well as for the specification of subcellular localization. Sirtuins are found in different cellular compartments: SIRT1 , SIRT6 , and SIRT7 are principally localized in the nucleus while other sirtuins reside in the mitochondria or in the cytoplasm as illustrated in Figure 1 b. SIRT7 is the only member of the family that is highly enriched in the nucleolus due to the presence of nuclear and nucleolar localization sequences at the N-terminal and C-terminal regions [7] ( Figure 1 b). Sirtuins efficiently shuttle between different compartments in response to numerous intracellular and extracellular stimuli to promote the activation of specific signaling pathways. These molecules activate a complex network of cellular responses to stress to maintain cellular homeostasis, including regulation of the transcription of specific target genes, modulation of chromatin structure, activation of mechanisms of DNA repair, metabolic adaptation to stress, and modulation of apoptosis among others [8] ( Figure 1 c). The ability of sirtuins to control such a broad range of biological functions mainly derives from their capacity to control both chromatin-related and -unrelated targets. Sirtuins act as potent epigenetic silencers by promoting heterochromatin formation through the direct deacetylation of different histone marks or indirectly by controlling the activity of numerous histone modifiers such as methytransferases and acetyltransferases [8,9][8][9]. Notwithstanding, sirtuins control the activity and functions of enzymes, transcription factors, and other chromatin unrelated molecules mainly through direct deacetylation, highlighting their complex functions in the regulation of cellular processes [3].
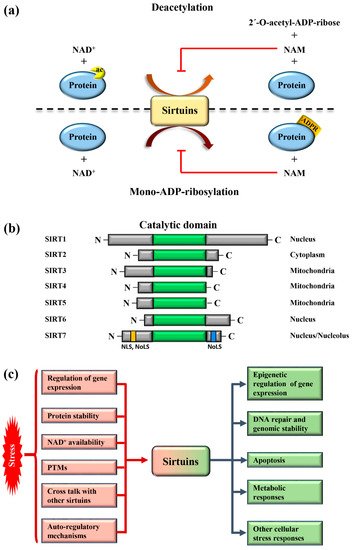
Different mechanisms are employed to promptly activate sirtuins following stress. Stressors modulate sirtuin genes’ expression, protein stability, or control their binding to specific inhibitors [5] ( Figure 1 c). Moreover, the strict dependency of the catalytic activity of sirtuins on NAD + allows swift adaption to the metabolic state of cells. Stress conditions such as glucose deprivation or caloric restriction (CR), which increase NAD + levels due to enhanced mitochondrial respiration and dramatically induce sirtuins expression and/or activity [6] ( Figure 1 c). CR represents the most prominent intervention to delay ageing and extend life span in different experimental organisms [15][10]. Different studies demonstrated that the beneficial effects of CR can be attributed to the activation of sirtuins, at least in part [16][11]. Thus, the activation of sirtuins might improve cellular adaptation to stress, induced by nutrient deprivation.
The acquisition of specific post-translational modifications (PTMs) such as phosphorylation, methylation, and ubiquitination, among others, represents another mechanism to control the activity of sirtuins. PTMs control the interaction of sirtuins with specific targets, their subcellular localization and catalytic activity, in some cases by interfering with the capacity to bind NAD + [17,18,19][12][13][14]. Intriguingly, recent studies also demonstrated that mammalian sirtuins possess auto-regulatory mechanisms. For instance, SIRT1 is capable of auto-catalytic activation by auto-deacetylation [20][15] while SIRT7 possesses auto-mono-ADP ribosylation activity [1]. Interestingly, cross-regulation between mammalian sirtuins is also employed to fine-tune their functions. The binding of SIRT7 to SIRT1 inhibits SIRT1 auto-catalytic activation resulting in the stimulation of adipogenesis and destabilization of constitutive heterochromatin [9,20][9][15]. In sharp contrast, binding of SIRT1 to SIRT7 is fundamental to promote the formation of ribosomal DNA heterochromatin and to repress E-cadherin expression, thus stimulating cancer metastasis [21,22][16][17]. Additionally, a synergistic effect between SIRT1 and SIRT6 has been described. SIRT1 -mediated deacetylation of SIRT6 is fundamental for recruitment of SIRT6 to double strand breaks (DSBs), thereby facilitating chromatin remodeling required for efficient DNA repair [23][18]. These findings illustrate the complexity of the regulation of sirtuins under physiological and pathological conditions. More research is warranted to unveil the molecular mechanisms controlling the regulation of these molecules in stress induced cellular processes, in order to identify novel pathways suitable for therapeutic interventions in diseases characterized by alterations in functions of sirtuins ( Figure 1 c).
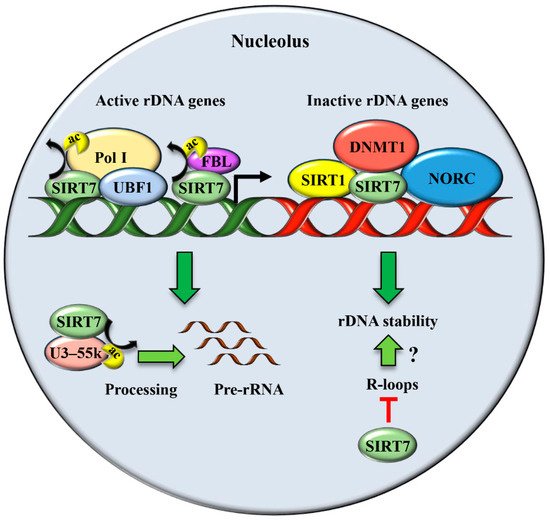
2. SIRT7 Controls Multiple Functions of the Nucleolus
| SIRT7 | Nucleolar Functions | ||||
|---|---|---|---|---|---|
| Function | Cell Type | Condition | |||
| Stimulation of rDNA transcription and ribosome biogenesis | Cancer cell lines and human embryonic kidney cells | Physiological conditions [27,28,]. | Physiological conditions [19] | 29, | [20] | 30, | [21][22] | 31 | [23]. |
| Maintenance of rDNA repeats integrity | Primary mouse embryonic fibroblasts (MEFs), mouse liver, human fibroblast-like fetal lung cells | Physiological conditions [22,47]. | Physiological conditions [17][27]. | ||||||
| Resolution of R-loops | Cancer cell lines and human embryonic kidney cells | Physiological conditions [53]. | Physiological conditions [28]. | ||||||
| Stabilization of p53 through NPM deacetylation | Cancer cell lines, MEFs, mouse skin | UV irradiation [17] | UV irradiation [12] | ||||||
| Stabilization of p53 through MDM2 degradation | Cancer cell lines | glucose starvation [59] | glucose starvation [29] | ||||||
| Inhibition of pre-rRNA transcription following stress | Cancer cell lines and human embryonic kidney cells | Nutrient stress [28,66]. hypertonic stress [31]. | Nutrient stress [20][30]. hypertonic stress [23]. |
Besides its well-characterized role in ribosomes biogenesis, the nucleolus has been recognized as a critical compartment involved in maintaining genomic stability, preventing ageing, and enabling cellular stress responses [37][31].
The translocation of SIRT7 in response to glucose starvation represents another mechanism to stabilize p53. Nucleoplasmic SIRT7 associates and deacetylates the acetyl transferase p300/CBP-associated factor (PCAF), which increases the binding of PCAF to MDM2 favoring MDM2 degradation, p53 stabilization, and cell cycle inhibition [59][29] ( Figure 3 and Table 1 ). Apparently, distinct mechanisms are employed by SIRT7 to inhibit MDM2 for promoting p53 stabilization, depending on the type of stress [17,59][12][29]. On the other hand, SIRT7 is not always required for p53 stabilization. For example, SIRT7 has no impact on p53 stabilization following inhibition of rDNA transcription [17][12] and rather promotes p53 stabilization in response to specific stress stimuli although through still poorly characterized mechanisms [60][32]. The capacity of SIRT7 to directly deacetylate p53 and suppress its transcriptional activity adds another layer of complexity into the mechanisms by which SIRT7 controls the p53 pathway. Although some studies demonstrated that the deacetylation of p53 by SIRT7 suppresses its pro-apoptotic functions and favors cell survival under stress [61[33][34],62], other studies failed to identify p53 as a deacetylation target of SIRT7 under particular stress conditions or in vitro [32,59,63][24][29][35]. These apparently contradictory results suggest a highly complex role of SIRT7 in controlling the p53 pathway that warrants further research.
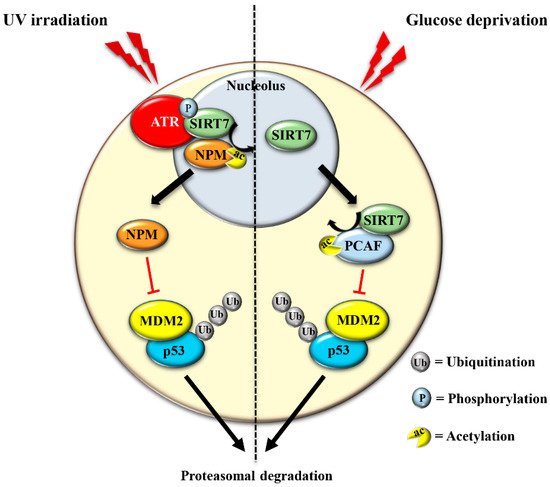
The exclusion of SIRT7 from nucleoli in response to different stress signals has been amply documented, indicating that exclusion of SIRT7 per se represents a hallmark of the NSR [36,65][36][37]. Several lines of evidence suggest a dynamic role of SIRT7 in the stressed nucleolus that might be employed to fine-tune activation of distinct cellular responses. In the early phase of the NSR, SIRT7 activates specific signaling pathways through deacetylation of nucleolar targets such as NPM [17][12]. The exclusion of SIRT7 from the nucleolus seems to represent a mechanism that prevents hyper-activation of nucleolar SIRT7 targets. Notwithstanding, the exit of SIRT7 from the nucleolus results in the hyperacetylation of other nucleolar proteins and consequent modulation of their functions, which may occur during a late phase of the NSR [36]. Furthermore, relocation of SIRT7 into other cellular compartments is instrumental for rapid activation of extra-nucleolar functions of SIRT7 .
The mechanisms controlling SIRT7 translocation from the nucleoli following stress stimuli remain poorly characterized. Maintenance of SIRT7 in the nucleoli is strictly correlated to rRNA transcription, since its inhibition excludes SIRT7 from this compartment [27][19]. Several different pathways inhibit rDNA transcription in response to stress. Thus, the reduction of rRNA expression itself may cause exclusion of SIRT7 from nucleoli, which will further reinforce transcriptional inhibition, robustly preventing ribosome biogenesis. Post-translational modifications also control the subcellular localization of SIRT7 . The phosphorylation of SIRT7 by AMPK (5’ adenosine monophosphate-activated protein kinase), a key molecule for restoring cellular energy levels, favors exclusion of SIRT7 from nucleoli, resulting in dramatic downregulation of rDNA transcription and energy saving following glucose starvation [66][30]. SIRT7 is also phosphorylated by the kinase ATR following genotoxic stress, although the effect of this post-translational modification on the subcellular distribution of SIRT7 and its contribution to rRNA transcription was not determined [17][12]. The complex network employed to efficiently exclude SIRT7 from the nucleolus has been only partially characterized, despite its paramount importance for inhibiting rDNA transcription and regulating other cellular functions ( Figure 4 and Table 1 ).
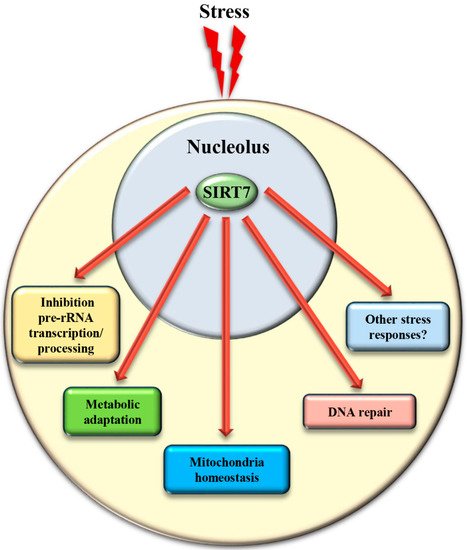
4. SIRT7 Controls Extra-Nucleolar Functions to Ensure Cellular Integrity Following Stress
In addition to the control of critical nucleolar functions, SIRT7 is involved in numerous cellular reactions that take place outside this compartment. We assume that the nucleolus acts as a reservoir of SIRT7 and ensures its rapid mobilization following stress to achieve a robust activation of SIRT7 -mediated extra-nucleolar functions. For instance, SIRT7 controls the activation of a transcriptional program that facilitates adaptation to starvation. This process requires auto-mono-ADP ribosylation of SIRT7 . Auto-modified SIRT7 binds to the histone variant mH2A1.1, which binds mono-ADP-ribose. Thereby, SIRT7 is recruited to intragenic regions where it controls expression of nearby genes by modulating chromatin organization [1]. SIRT7 is also involved in maintaining mitochondria homeostasis. Mitochondria are organelles responsible for cellular energy generation through the biosynthesis of ATP but also control other cellular activities. Dysregulation of mitochondrial homeostasis has been implicated in the development of numerous human diseases including ageing. Moreover, the adaptation of mitochondrial functions in response to nutrient deprivation is imperative for the maintenance of cellular homeostasis and to ensure cell survival [67][38]. Ablation of SIRT7 in mice correlates with the onset of cellular and organismal alterations associated with mitochondrial dysfunctions such as accelerated ageing, deterioration of hematopoietic stem cells, cardiac and hepatic diseases, and age-associated hearing loss [68,69][39][40]. SIRT7 employs different mechanisms to control mitochondrial functions. In particular, SIRT7 acts as a potent epigenetic suppressor of nuclear encoded genes responsible for mitochondria biogenesis [69,70][40][41] and represses thereby the mitochondrial protein folding stress response (PFS mt ). Inhibition of SIRT7 enzymatic activity under physiological conditions through PRMT6-mediated methylation ensures efficient stimulation of mitochondria biogenesis [70][41]. Following glucose starvation, binding of SIRT7 to PRMT6 is disrupted via a mechanism requiring AMPK-dependent phosphorylation, which stimulates SIRT7 -mediated epigenetic repression of target genes leading to inhibition of de novo biosynthesis of mitochondria [70][41]. In addition, SIRT7 -dependent inhibition of the PFS mt ensures cell survival following nutrient deprivation [69][40]. On the other hand, SIRT7 stimulates expression of genes controlling mitochondrial functions (such as components of the respiratory chain) by activating the transcription factor GABPβ1 through direct deacetylation [68][39].
References
- Simonet, N.G.; Thackray, J.K.; Vazquez, B.N.; Ianni, A.; Espinosa-Alcantud, M.; Morales-Sanfrutos, J.; Hurtado-Bagès, S.; Sabidó, E.; Buschbeck, M.; Tischfield, J.; et al. SirT7 auto-ADP-ribosylation regulates glucose starvation response through mH2A1. Sci. Adv. 2020, 6, eaaz2590.
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Choi, B.H.; He, B.; et al. Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 2011, 334, 806–809.
- Ianni, A.; Yuan, X.; Bober, E.; Braun, T. Sirtuins in the Cardiovascular System: Potential Targets in Pediatric Cardiology. Pediatr. Cardiol. 2018, 39, 983–992.
- Li, L.; Shi, L.; Yang, S.; Yan, R.; Zhang, D.; Yang, J.; He, L.; Li, W.; Yi, X.; Sun, L.; et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 2016, 7, 12235.
- Feldman, J.L.; Dittenhafer-Reed, K.; Denu, J.M. Sirtuin Catalysis and Regulation. J. Biol. Chem. 2012, 287, 42419–42427.
- Imai, S.-I.; Guarente, L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. Npj Aging Mech. Dis. 2016, 2, 16017.
- Kiran, S.; Chatterjee, N.; Singh, S.; Kaul, S.; Wadhwa, R.; Ramakrishna, G. Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J. 2013, 280, 3451–3466.
- Bosch-Presegué, L.; Vaquero, A. Sirtuins in stress response: Guardians of the genome. Oncogene 2013, 33, 3764–3775.
- Kumari, P.; Popescu, D.; Yue, S.; Bober, E.; Ianni, A.; Braun, T. Sirt7 inhibits Sirt1-mediated activation of Suv39h1. Cell Cycle 2018, 17, 1403–1412.
- Anderson, R.M.; Weindruch, R. The caloric restriction paradigm: Implications for healthy human aging. Am. J. Hum. Biol. 2012, 24, 101–106.
- Guarente, L.P. Calorie restriction and sirtuins revisited. Genes Dev. 2013, 27, 2072–2085.
- Ianni, A.; Kumari, P.; Tarighi, S.; Simonet, N.G.; Popescu, D.; Guenther, S.; Hölper, S.; Schmidt, A.; Smolka, C.; Yue, S.; et al. SIRT7-dependent deacetylation of NPM promotes p53 stabilization following UV-induced genotoxic stress. Proc. Natl. Acad. Sci. USA 2021, 118, e2015339118.
- Flick, F.; Lüscher, B. Regulation of Sirtuin Function by Posttranslational Modifications. Front. Pharmacol. 2012, 3, 29.
- Kalous, K.S.; Wynia-Smith, S.; Olp, M.D.; Smith, B.C. Mechanism of Sirt1 NAD+-dependent Protein Deacetylase Inhibition by Cysteine S-Nitrosation. J. Biol. Chem. 2016, 291, 25398–25410.
- Fang, J.; Ianni, A.; Smolka, C.; Vakhrusheva, O.; Nolte, H.; Krüger, M.; Wietelmann, A.; Simonet, N.; Adrian-Segarra, J.M.; Vaquero, A.; et al. Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc. Natl. Acad. Sci. USA 2017, 114, E8352–E8361.
- Malik, S.; Villanova, L.; Tanaka, S.; Aonuma, M.; Roy, N.; Berber, E.; Pollack, J.R.; Michishita-Kioi, E.; Chua, K.F. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci. Rep. 2015, 5, 9841.
- Ianni, A.; Hoelper, S.; Krueger, M.; Braun, T.; Bober, E. Sirt7 stabilizes rDNA heterochromatin through recruitment of DNMT1 and Sirt1. Biochem. Biophys. Res. Commun. 2017, 492, 434–440.
- Meng, F.; Qian, M.; Peng, B.; Peng, L.; Wang, X.; Zheng, K.; Liu, Z.; Tang, X.; Zhang, S.; Sun, S.; et al. Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. eLife 2020, 9, e55828.
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–1080.
- Chen, S.; Seiler, J.; Santiago-Reichelt, M.; Felbel, K.; Grummt, I.; Voit, R. Repression of RNA Polymerase I upon Stress Is Caused by Inhibition of RNA-Dependent Deacetylation of PAF53 by SIRT7. Mol. Cell 2013, 52, 303–313.
- Iyer-Bierhoff, A.; Krogh, N.; Tessarz, P.; Ruppert, T.; Nielsen, H.; Grummt, I. SIRT7-Dependent Deacetylation of Fibrillarin Controls Histone H2A Methylation and rRNA Synthesis during the Cell Cycle. Cell Rep. 2018, 25, 2946–2954.
- Grob, A.; Roussel, P.; Wright, J.E.; McStay, B.; Hernandez-Verdun, D.; Sirri, V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 2009, 122, 489–498.
- Chen, S.; Blank, M.F.; Iyer, A.; Huang, B.; Wang, L.; Grummt, I.; Voit, R. SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat. Commun. 2016, 7, 10734.
- Barber, M.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.; Chen, K.; et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nat. Cell Biol. 2012, 487, 114–118.
- Pandey, V.; Kumar, V. Stabilization of SIRT7 deacetylase by viral oncoprotein HBx leads to inhibition of growth restrictive RPS7 gene and facilitates cellular transformation. Sci. Rep. 2015, 5, 14806.
- Tsai, Y.-C.; Greco, T.; Cristea, I.M. Sirtuin 7 Plays a Role in Ribosome Biogenesis and Protein Synthesis. Mol. Cell. Proteom. 2014, 13, 73–83.
- Paredes, S.; Angulo-Ibanez, M.; Tasselli, L.; Carlson, S.M.; Zheng, W.; Li, T.-M.; Chua, K.F. The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. J. Biol. Chem. 2018, 293, 11242–11250.
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017, 31, 1370–1381.
- Lu, Y.-F.; Xu, X.-P.; Zhu, Q.; Liu, G.; Bao, Y.-T.; Wen, H.; Li, Y.-L.; Gu, W.; Zhu, W.-G. SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene 2020, 39, 4650–4665.
- Sun, L.; Fan, G.; Shan, P.; Qiu, X.; Dong, S.; Liao, L.; Yu, C.; Wang, T.; Gu, X.; Li, Q.; et al. Regulation of energy homeostasis by the ubiquitin-independent REGγ proteasome. Nat. Commun. 2016, 7, 12497.
- Stochaj, U.; Weber, S.C. Nucleolar Organization and Functions in Health and Disease. Cells 2020, 9, 526.
- Kumari, P.; Tarighi, S.; Braun, T.; Ianni, A. The complex role of SIRT7 in p53 stabilization: Nucleophosmin joins the debate. Mol. Cell. Oncol. 2021, 8, 1896349.
- Sun, M.; Zhai, M.; Zhang, N.; Wang, R.; Liang, H.; Han, Q.; Jia, Y.; Jiao, L. MicroRNA-148b-3p is involved in regulating hypoxia/reoxygenation-induced injury of cardiomyocytes in vitro through modulating SIRT7/p53 signaling. Chem. Interact. 2018, 296, 211–219.
- Zhao, J.; Wozniak, A.; Adams, A.; Cox, J.; Vittal, A.; Voss, J.; Bridges, B.; Weinman, S.A.; Li, Z. SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J. Exp. Clin. Cancer Res. 2019, 38, 252.
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Mol. Biol. Cell 2005, 16, 4623–4635.
- Blank, M.F.; Grummt, I. The seven faces of SIRT7. Transcription 2017, 8, 67–74.
- Kiran, S.; Oddi, V.; Ramakrishna, G. Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp. Cell Res. 2015, 331, 123–141.
- Liesa, M.; Shirihai, O.S. Mitochondrial Dynamics in the Regulation of Nutrient Utilization and Energy Expenditure. Cell Metab. 2013, 17, 491–506.
- Ryu, D.; Jo, Y.S.; Lo Sasso, G.; Stein, S.; Zhang, H.; Perino, A.; Lee, J.U.; Zeviani, M.; Romand, R.; Hottiger, M.O.; et al. A SIRT7-Dependent Acetylation Switch of GABPβ1 Controls Mitochondrial Function. Cell Metab. 2014, 20, 856–869.
- Mohrin, M.; Shin, J.; Liu, Y.; Brown, K.; Luo, H.; Xi, Y.; Haynes, C.M.; Chen, D. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015, 347, 1374–1377.
- Yan, W.-W.; Liang, Y.-L.; Zhang, Q.-X.; Wang, D.; Lei, M.-Z.; Qu, J.; He, X.-H.; Lei, Q.-Y.; Wang, Y.-P. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 2018, 19, e46377.
