The R2R3-MYB transcription factors (TFs) play several key roles in numerous plant biological processes. Hedychium coronarium is an important ornamental plant well-known for its elegant flower shape and abundant aroma type. The floral aroma of H. coronarium is due to the presence of a large amount of terpenes and benzenoids. However, less is known about the role of R2R3-MYB TFs in the regulatory mechanism of floral aroma production in this breed. Herein, we isolate and functionally characterize the R2R3-MYB TF HcMYB132, which is potentially involved in regulating floral aroma synthesis. Sequence alignment analysis revealed that it includes a nuclear localization signal NLS(s) and a 2R, 3R motif signature in the sequences. A subcellular localization assay revealed that HcMYB132 protein localizes to the nucleus. Real-time qPCR assays showed that HcMYB132 is specifically expressed in flowers and its expression pattern correlates with the emission of floral volatile compounds. In HcMYB132-silenced flowers, the levels of floral volatile compounds were significantly reduced, and the expression of key structural volatile synthesis genes was downregulated compared to control. Collectively, these results suggest that HcMYB132 might play a significant role in the regulation of terpenoid biosynthesis in H. coronarium.
- floral scent
- Hedychium coronarium
- R2R3-MYB
- structural genes
- terpenes
1. Introduction
2. Characterization of HcMYB132
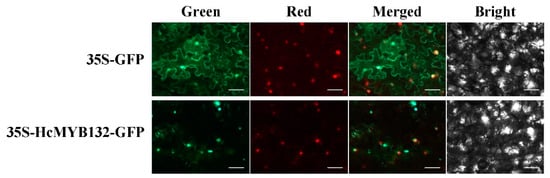
3. Subcellular Localization of HcMYB132
2.3. Expression Pattern of HcMYB132


4. Discussion
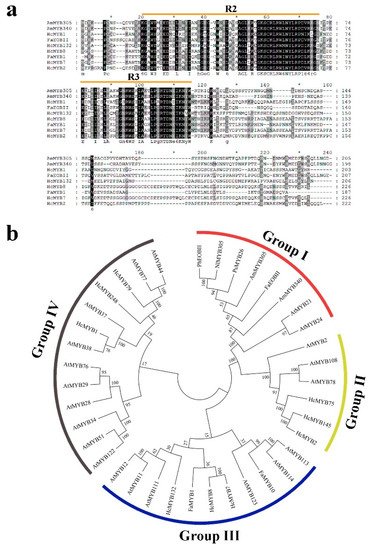
H. coronarium is popular in tropical and subtropical parts of the world due to its appealing strong aroma type and medicinal properties [3,33][3][33]. R2R3-MYB TFs are the main regulators of terpenes and phenylpropanoids [34,35][34][35]. However, less is known about the transcriptional regulatory mechanism of floral aroma production. Until now, a few MYB TFs have been reported that control the regulatory network of floral scent production [29,30,36,37][29][30][36][37]. Herein, we identified and functionally characterized a R2R3-MYB TF (HcMYB132) that is potentially involved in floral aroma synthesis in H. coronarium. Multiple sequence analyses of HcMYB132 revealed the existence of 2R and 3R repeats in the sequences (Figure 1a). Several previous findings suggest that the R2 and R3 signature motifs are highly conserved and regulate various aspects of plant secondary metabolites [13,38,39,40][13][38][39][40]. We generated a phylogenic tree using the previously characterized R2R3-MYB TFs involved in the regulatory network of secondary metabolism, together with HcMYB132 (Figure 1b). HcMYB132 was classified into Group III with FaMYB1, FaMYB10, and AtMYB11/12/111/113/114/123. The functional characterization of aforementioned genes revealed their role in the regulation of the flavonoid/phenylpropanoid metabolism [14,41[14][41][42][43],42,43], indicating that HcMYB132 might play a significant role in secondary metabolism. It has been reported that MYB TFs in same subclade have identical functions [13,35][13][35]. The structure analysis revealed that the HcMYB132 contains two exons, which are in line with the previous reports [44]. A subcellular localization assay revealed that HcMYB132 protein is localized to the nucleus, which is consistent with the previous findings [1,7,13,45][1][7][13][45]. The process of floral scent production is interrelated with flower development [46,47,48][46][47][48]. Our previous studies revealed that production and emission of floral volatile compounds and the expression of key structural volatile biosynthesis genes were low during the bud stage and peaked during the full bloom stage [7,8,9,10][7][8][9][10]. Previous studies also showed that volatile emission content was significantly larger from the flower than from the rhizome and leaf, which is consistent with the expression pattern of HcMYB132 [7]. In the current findings, it was revealed that HcMYB132 was mainly expressed in the flowers and its expression pattern increased with flower development, peaked during the fully bloomed stage, and dropped down thereafter (Figure 4a,b), implying that it might potentially be involved in the floral aroma production and emission mechanism. A similar expression pattern was observed in Fragaria ananassa EOBII, EOBI, and ODO1, and was involved in the regulatory network of eugenol [15,29][15][29]. Likewise, Prunus persica MYBF1 and MYB15 showed the highest expression in the flower and were involved in flavanol biosynthesis regulation [31]. In Lilium hybrid, ODO1 TF had highest expression in the flower and plaedy a crucial role in the regulation of phenylpropanoid/ benzenoid volatile production [49]. These results suggest that HcMYB132 potentially regulates the process of floral scent production. To reveal the role of HcMYB132 in floral aroma production in H. coronarium, the activity of HcMYB132 was repressed via gene silencing. The data showed that the volatile contents of eucalyptol were substantially decreased in HcMYB132-silenced flowers compared to control flowers. Furthermore, in HcMYB132-silenced flowers, the transcript levels of key eucalyptol volatile biosynthesis genes (HcTPS1 and HcTPS3) were significantly decreased (Figure 5). Likewise, strawberry MYB10 regulates the expression of numerous key genes involved in the flavonoid and phenylpropanoid biosynthesis process [14]. In petunia ODO1-suppressed plants, the mRNA levels of several scent-related genes were downregulated [29]. Similarly, litchi MYB5 activates the transcript levels of key genes involved in the synthesis of anthocyanin [23]. In HcMYB1/2/7/8/75/79/145/238/248-silenced flowers, the emission of floral volatiles and the expression of structural genes were significantly decreased [1,7][1][7]. Moreover, the emission of eucalyptol and the expression of HcMYB132 were influenced by auxin treatments, which are consistent with previous findings [7,50][7][50]. These data endorse the previous findings that R2R3-MYB TFs are involved in the regulation of volatile formation in H. coronarium.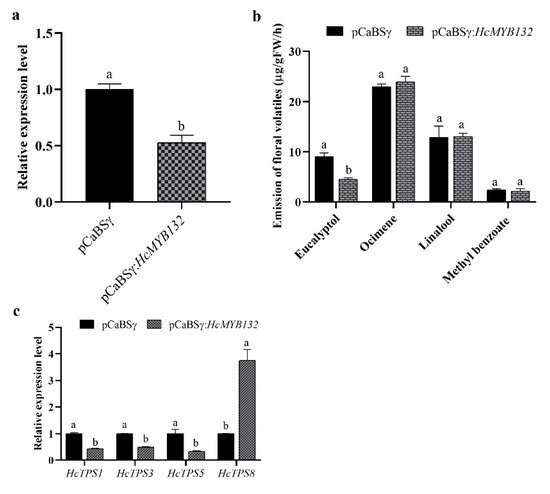
References
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, Y.; Waseem, M.; Ashraf, D.; Wang, C.; Wang, X.; Li, X.; Yue, Y. Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 2021, 12, 58.
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, Y.; Waseem, M.; Ashraf, U.; Li, X.; Yu, R.; Fan, Y. Genome-wide analysis of ARF transcription factors reveals HcARF5 expression profile associated with the biosynthesis of β-ocimene synthase in Hedychium coronarium. Plant Cell Rep. 2021, 40, 1269–1284.
- Báez, D.; Pino, J.A.; Morales, D. Floral scent composition in Hedychium coronarium J. Koenig analyzed by SPME. J. Essen. Oil Res. 2011, 23, 64–67.
- Fan, Y.; Yu, R.; Huang, Y.; Chen, Y. Studies on the essential constituent of Hedychium flavum and H. coronarium. Acta Hortic. Sin. 2003, 30, 475.
- Fan, Y.-p.; Wang, X.-r.; Yu, R.-c.; Yang, P. Analysis on the aroma components in several species of Hedychium. Acta Hortic. Sin. 2007, 34, 231.
- Wang, C.; Abbas, F.; Zhou, Y.; Ke, Y.; Li, X.; Yue, Y.; Yu, Y.; Yu, R.; Fan, Y. Genome-wide identification and expression pattern of SnRK gene family under several hormone treatments and its role in floral scent emission in Hedychium coronarium. PeerJ 2021, 9, e10883.
- Ke, Y.; Abbas, F.; Zhou, Y.; Yu, R.; Fan, Y. Auxin-responsive R2R3-MYB transcription factors HcMYB1 and HcMYB2 activate volatile biosynthesis in Hedychium coronarium flowers. Front. Plant Sci. 2021, 12, 1626.
- Ke, Y.; Abbas, F.; Zhou, Y.; Yu, R.; Yue, Y.; Li, X.; Yu, Y.; Fan, Y. Genome-wide analysis and characterization of the Aux/IAA Family genes related to floral scent formation in Hedychium coronarium. Int. J. Mol. Sci. 2019, 20, 3235.
- Yue, Y.; Yu, R.; Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762.
- Yue, Y.; Yu, R.; Fan, Y. Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genom. 2015, 16, 470.
- Ramya, M.; Kwon, O.K.; An, H.R.; Park, P.M.; Baek, Y.S.; Park, P.H. Floral scent: Regulation and role of MYB transcription factors. Phytochem. Lett. 2017, 19, 114–120.
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953.
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581.
- Medina-Puche, L.; Cumplido-Laso, G.; Amil-Ruiz, F.; Hoffmann, T.; Ring, L.; Rodríguez-Franco, A.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J.; Blanco-Portales, R. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 2014, 65, 401–417.
- Medina-Puche, L.; Molina-Hidalgo, F.J.; Boersma, M.; Schuurink, R.C.; López-Vidriero, I.; Solano, R.; Franco-Zorrilla, J.-M.; Caballero, J.L.; Blanco-Portales, R.; Muñoz-Blanco, J. An R2R3-MYB transcription factor regulates eugenol production in ripe strawberry fruit receptacles. Plant Physiol. 2015, 168, 598–614.
- Chen, K.; Du, L.; Liu, H.; Liu, Y. A novel R2R3-MYB from grape hyacinth, MaMybA, which is different from MaAN2, confers intense and magenta anthocyanin pigmentation in tobacco. BMC Plant Biol. 2019, 19, 390.
- Chen, R.; Ni, Z.; Nie, X.; Qin, Y.; Dong, G.; Sun, Q. Isolation and characterization of genes encoding Myb transcription factor in wheat (Triticum aestivem L.). Plant Sci. 2005, 169, 1146–1154.
- Chen, T.; Li, W.; Hu, X.; Guo, J.; Liu, A.; Zhang, B. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015, 56, 917–929.
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Do Choi, Y.; Cheong, J.-J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635.
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721.
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176.
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161.
- Lai, B.; Du, L.-N.; Hu, B.; Wang, D.; Huang, X.-M.; Zhao, J.-T.; Wang, H.-C.; Hu, G.-b. Characterization of a novel litchi R2R3-MYB transcription factor that involves in anthocyanin biosynthesis and tissue acidification. BMC Plant Biol. 2019, 19, 62.
- Pérez-Díaz, J.R.; Pérez-Díaz, J.; Madrid-Espinoza, J.; González-Villanueva, E.; Moreno, Y.; Ruiz-Lara, S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016, 90, 63–76.
- Yan, J.; Wang, B.; Zhong, Y.; Yao, L.; Cheng, L.; Wu, T. The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 2015, 89, 35–48.
- Shin, B.; Choi, G.; Yi, H.; Yang, S.; Cho, I.; Kim, J.; Lee, S.; Paek, N.C.; Kim, J.H.; Song, P.S. AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 2002, 30, 23–32.
- Uimari, A.; Strommer, J. Myb26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 1997, 12, 1273–1284.
- Verdonk, J.C.; Haring, M.A.; van Tunen, A.J.; Schuurink, R.C. ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 2005, 17, 1612–1624.
- Spitzer-Rimon, B.; Farhi, M.; Albo, B.; Cna’ani, A.; Zvi, M.M.B.; Masci, T.; Edelbaum, O.; Yu, Y.; Shklarman, E.; Ovadis, M. The R2R3-MYB–like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 2012, 24, 5089–5105.
- Spitzer-Rimon, B.; Marhevka, E.; Barkai, O.; Marton, I.; Edelbaum, O.; Masci, T.; Prathapani, N.-K.; Shklarman, E.; Ovadis, M.; Vainstein, A. EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. Plant Cell 2010, 22, 1961–1976.
- Cao, Y.; Xie, L.; Ma, Y.; Ren, C.; Xing, M.; Fu, Z.; Wu, X.; Yin, X.; Xu, C.; Li, X. PpMYB15 and PpMYBF1 transcription factors are involved in regulating flavonol biosynthesis in peach fruit. J. Agric. Food Chem. 2018, 67, 644–652.
- El-Azaz, J.; de la Torre, F.; Pascual, M.B.; Debille, S.; Canlet, F.; Harvengt, L.; Trontin, J.-F.; Ávila, C.; Cánovas, F.M. Transcriptional analysis of arogenate dehydratase genes identifies a link between phenylalanine biosynthesis and lignin biosynthesis. J. Exp. Bot. 2020, 71, 3080–3093.
- Wu, Z.; Raven, P. Flora of China. Vol. 24 (Flagellariaceae through Marantaceae); Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2000.
- An, X.-H.; Tian, Y.; Chen, K.-Q.; Liu, X.-J.; Liu, D.-D.; Xie, X.-B.; Cheng, C.-G.; Cong, P.-H.; Hao, Y.-J. MdMYB9 and MdMYB11 are Involved in the Regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cell Physiol. 2014, 56, 650–662.
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.-X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156.
- Bedon, F.; Bomal, C.; Caron, S.; Levasseur, C.; Boyle, B.; Mansfield, S.D.; Schmidt, A.; Gershenzon, J.; Grima-Pettenati, J.; Séguin, A. Subgroup 4 R2R3-MYBs in conifer trees: Gene family expansion and contribution to the isoprenoid-and flavonoid-oriented responses. J. Exp. Bot. 2010, 61, 3847–3864.
- Van Moerkercke, A.; Haring, M.A.; Schuurink, R.C. The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. Plant J. 2011, 67, 917–928.
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677.
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456.
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB transcription factors–Functions outside the DNA-binding domain. Trends Plant Sci. 2019, 24, 934–946.
- Aharoni, A.; de Vos, C.R.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332.
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708.
- Pandey, A.; Misra, P.; Bhambhani, S.; Bhatia, C.; Trivedi, P.K. Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 2014, 4, 5018.
- Liu, C.; Hao, J.; Qiu, M.; Pan, J.; He, Y. Genome-wide identification and expression analysis of the MYB transcription factor in Japanese plum (Prunus salicina). Genomics 2020, 112, 4875–4886.
- Colquhoun, T.A.; Kim, J.Y.; Wedde, A.E.; Levin, L.A.; Schmitt, K.C.; Schuurink, R.C.; Clark, D.G. PhMYB4 fine-tunes the floral volatile signature of Petunia× hybrida through PhC4H. J. Exp. Bot. 2010, 62, 1133–1143.
- Abbas, F.; Ke, Y.; Yu, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 2017, 246, 803–816.
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32.
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Fäldt, J.; Miller, B.; Bohlmann, J. (E)-β-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 2003, 15, 1227–1241.
- Yoshida, K.; Oyama-Okubo, N.; Yamagishi, M. An R2R3-MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp.). Mol. Breed. 2018, 38, 144.
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453.
