Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Andrés J. Cortés.
Warming and drought are reducing global crop production with a potential to substantially worsen global malnutrition. As with the green revolution in the last century, plant genetics may offer concrete opportunities to increase yield and crop adaptability. However, the rate at which the threat is happening requires powering new strategies in order to meet the global food demand.
- abiotic stress tolerance
- genebanks
- germplasm collections
- ex situ conservation
- genetic adaptation
- genome-wide selection scans (GWSS)
- genome–environment associations (GEA)
- genomic prediction (GP)
- machine learning (ML)
1. Introduction—The Imminence of the Threat
How plants and crops will respond to a warmer and drier climate is currently one of the most discussed multi-disciplinary questions in the fields of environmental science, ecology, and evolution. It is estimated that climate change effects may limit global crop production by at least 10% in 2050 [1[1][2],2], especially in vulnerable regions around the globe where heat, drought, and malnutrition are already substantial. Given that current agricultural resources might not be sufficient to meet future food demand [3], crop wild relatives and landraces historically adapted to dry and semi-arid environments are key sources of yet unexplored diversity with respect to major food crops [4]. Exotics may donate necessary genetic variation to make heat and drought-tolerant cultivars, or may even stand as novel crops by themselves [5,6,7][5][6][7] (e.g., Lupinus mutabilis [8]).
However, efforts to capture and pyramid target tolerant variants from the wild still face major challenges. First, identifying useful variation in wild accessions through field trials (Figure 1a) has been inefficient because of the disparity in growth rates and phenologies [9]. This limitation is reinforced by a complex inheritance of the abiotic stress tolerant phenotypes involving many genes of low effects and several environmental interactions [10]. Second, the domestication syndrome typically neglected heat and drought tolerance in the majority of crop species [11,12][11][12] because these adaptations in wild populations (Figure 1b) tend to perpetuate vegetative phases, delay reproductive stages, and therefore compromise overall yield [13]. Due to this, transferring variation from wild exotic donors into elite lines may induce undesired linked trait variation such as flowering delay and reduced crop yields (e.g., in Phaseolus species [14]).
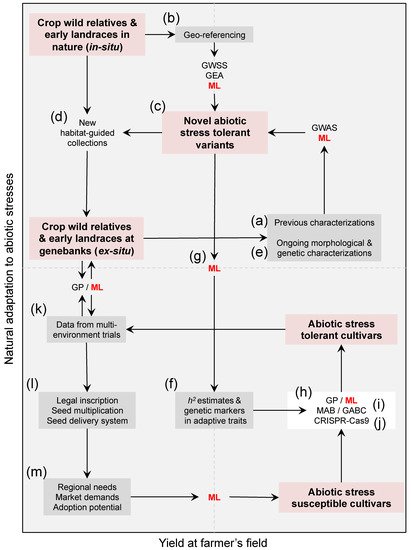
Figure 1. A roadmap of trans-disciplinary approaches aiming at harnessing genebank utilization for climate change research in the face of heat, and water scarcity. Compiling (a) previous characterizations and (b) geo-referencing-derived climate data/indices of available genetic resources in genebanks is a starting point to (c) assess the extent of abiotic stress tolerance among existing accessions, and the need of (d) new habitat-based population-guided collections targeting isolated pockets of cryptic diversity in dry and semi-arid regions. Planning question-oriented collecting trips of crop wild relatives and hidden landraces across contrasting environments/agro-ecologies is needed now more than ever, despite a century of gathering and preserving diversity in plants throughout genebanks. Coupling ex situ agro-ecological screenings together with (e) ongoing in situ genebanks characterizations for morphological and genetic variation is essential to define (c) putative tolerant reference collections, while understanding the (f) heritability (h2) of adaptive traits and their genetic architecture (i.e., underlying genes) via genome-wide selection scans (GWSS), genome–environment associations (GEA), and genome-wide association studies (GWAS). Since identifying these novel sources of heat and drought tolerance demands merging heterogeneous datasets, (g) machine learning (ML, in red letters) promises speeding up genebank characterization. The distinction that clustering (Table 1) and ML (Figure 2 and Table 2) strategies can provide between abiotic stress tolerant and susceptible accessions is essential to (h) transfer useful genetic variation from wild crop donors and early landraces into elite cultivated lines, either by designing (i) genomic-assisted breeding programs such as genomic prediction (GP) and inter-specific marker- and genomic-assisted backcrossing (MAB and GABC) schemes, or by envisioning (j) multi-trait gene editing strategies (e.g., CRISPR-Cas9). Once (k) abiotic stress tolerant varieties are validated across different environments, (l) legal inscription, seed multiplication, seed delivery system to farmers’ associations, and (m) follow-up given the regional needs, market demands, and adoption potential, are necessary downstream validation steps. These heterogeneous datasets are also likely to be inputted into ML, and in turn feedback new needs beyond heat and drought tolerance such as other types of resistances and nutritional quality. For ML to succeed speeding up the breeding of heat and drought-tolerant crops, there must be long-term funding to generate and maintain an assortment of datasets at each step, which in turn need to be publicly available through open access repositories from various geographic locations. Red boxes highlight different reservoirs of wild and cultivated diversity within the Cartesian space, gray boxes are mixed datasets built around these collections, and connectors are methodological approaches.
With the advent of the genomic era, heat and drought tolerance molecular pathways have been extensively studied [15[15][16][17],16,17], and numerous candidate genes and genetic marker associations have already been identified and validated (e.g., ABA-dependent or independent pathways, and ASR, DREB, and ERECTA-encoding genes [18,19,20,21][18][19][20][21]). Coupling these comprehensive datasets with novel analytical tools harbors the potential to identify and unlock useful genetic variation among crop wild relatives and landraces to challenge with abiotic stresses. In the following sections, we outline the main avenues to establish novel sources of abiotic stress tolerance variation from wild crop relatives and semi-domesticated landraces (Figure 1c), and discuss modern genomic-assisted strategies to utilize these variants to obtain heat and drought-tolerant elite crop lines. We finish by emphasizing the need to enable the construction and long-term maintenance of big heterogeneous dataset repositories capable of powering these innovative predictive strategies.
2. Valuing the Wild—Strategies to Identify Naturally Available Exotic Variants
Domestication has been the most ambitious evolutionary experiment humanity has ever embarked on [11,22][11][22]. Being the longest running selection trial [23], it has served as a playground for geneticists and biologists to explore the patterns and processes during crop evolution [24,25,26][24][25][26]. For instance, it has allowed humans to test hypothesis such as whether dual domestication syndromes have recruited the same genetic variants in parallel [27], or whether genomic divergence is more prone to harbor signatures of selection due to reduced recombination and increased drift [28]. In this way, domestication has invited researchers to study the repeatability of evolution [29], and the relative role of isolation, migration, and hybridization [30], long standing questions in evolutionary biology [31,32,33][31][32][33]. Similarly, studying crop evolution has proven enlightening, and addressing their wild relatives’ natural adaptation to distinct habitats (Figure 1b) informs on how plant phenotypes may react to a changing climate [34]. After all, natural selection has already tested more options than humans ever will [35].
2.1. On the Necessity of Broadening the Germplasm
Natural selection rarely misses key adaptive improvements. For instance, it has already improved the efficiency of photosynthesis and water use [35], which are major developments to enhance the yield potential that has increased little in recent decades. Therefore, inferring in situ genetic adaptation to heat and drought stresses, usually from ex situ geo-referenced widespread collections of crop wild relatives, has become a prerequisite to capture these naturally available exotic variants. Unfortunately, after a century of collecting and preserving diversity in plants throughout germplasm banks, living seed collections that serve as repositories of genetic and ecological variation [36], ex situ collections do not necessarily fully span the ecological niche of agricultural crop relatives [37,38][37][38] as to establish new sources of genes for improving complex adaptive traits. Nowadays, wild relatives are still discovered [39], and further expeditions (Figure 1d) are needed (e.g., in the ecological hyper-variable and species-rich neotropics) [40,41][40][41]. Because isolated pockets of cryptic diversity still persist, novel habitat-based population-guided collections for genebanks are paramount, now more than ever.
2.2. Going Global by Adapting Local
Germplasm collections should be used not only to introduce exotic variation but also to avoid genetic erosion and increase long-term adaptability to climate change by making crops more resistant to abiotic stresses such as heat and drought. Local adaptation to abiotic factors can be studied using historical climate at the habitats where geo-referenced germplasm accessions were originally collected (Figure 1b). If an ecological balance [42] between genotypes and environments can be assumed [43,44][43][44] (Humboldt’s ‘harmony in nature’), geo-referencing and repositories of in situ climate variables can then be used to compute adaptive capacity. The former condition typically applies for landraces and crop wild relatives because they have occupied local niches long enough as to be shaped by natural selection forces [45]. This approach has typically relied on clustering algorithms to predict (Table 1) not only drought tolerance, water use efficiency [46[46][47],47], and thermal tolerance [48], but also resistance to pathogens [49], and aluminum toxicity [50] in a wide range of landraces and wild species.
Genomic analytical tools (Figure 1e) commonly coupled with environmental variables in order to reconstruct the genetic architecture (Figure 1f) of adaptive trait variation to abiotic stresses are genome-wide selection scans (GWSS) [51] and genome–environment associations (GEA) [52]. Both approaches aim to capture the signatures of selection to different environments by retrieving those genomic regions that segregate and are fixed among contrasting habitats (e.g., arid vs. wet regions) [53,54][53][54]. The strategies differ in that the former uses outlier tests given a background (‘baseline’) distribution, usually within a Bayesian framework [55], while the latter relies on mixed linear models (MLMs) that explicitly incorporate covariates as random effects [56]. Because these analyses may be misleading [57[57][58],58], if confusing factors [33,59][33][59] are not appropriately accounted for, MLMs are nowadays the preferred method to describe the genetic basis of local adaptation in germplasm collections [60]. Other major improvements are the use of indices, rather than raw environmental variables, that summarize precise physiological processes (e.g., thermal thresholds and potential evapotranspiration models to infer heat/drought stress) [10[10][18][19][20],18,19,20], and the collection of spatial high-resolution climate data to make accurate predictions at the regional [61] and micro-habitat [62,63][62][63] levels.
References
- Tai, A.P.K.; Martin, M.V.; Heald, C.L. Threat to future global food security from climate change and ozone air pollution. Nat. Clim. Chang. 2014, 4, 817–821.
- Davis, K.F.; Gephart, J.A.; Emery, K.A.; Leach, A.M.; Galloway, J.N.; D’Odorico, P. Meeting future food demand with current agricultural resources. Glob. Environ. Chang. 2016, 39, 125–132.
- McCouch, S. Feeding the future. Nature 2013, 499, 23–24.
- Blair, M.W.; Pantoja, W.; Carmenza Munoz, L. First use of microsatellite markers in a large collection of cultivated and wild accessions of tepary bean (Phaseolus acutifolius A. Gray). Appl. Genet. 2012, 125, 1137–1147.
- Borelli, T.; Hunter, D.; Powell, B.; Ulian, T.; Mattana, E.; Termote, C.; Pawera, L.; Beltrame, D.; Penafiel, D.; Tan, A.; et al. Born to Eat Wild: An Integrated Conservation Approach to Secure Wild Food Plants for Food Security and Nutrition. Plants 2020, 9, 1299.
- von Wettberg, E.; Davis, T.M.; Smýkal, P. Wild Plants as Source of New Crops. Front. Plant. Sci. 2020, 11.
- Gulisano, A.; Alves, S.; Martins, J.N.; Trindade, L.M. Genetics and Breeding of Lupinus mutabilis: An Emerging Protein Crop. Front. Plant. Sci. 2019, 10, 1385.
- Beebe, S.E.; Rao, I.M.; Blair, M.W.; Acosta-Gallegos, J.A. Phenotyping common beans for adaptation to drought. Front. Physiol. 2013, 4, 35.
- Cortés, A.J.; Blair, M.W. Genotyping by Sequencing and Genome—Environment Associations in Wild Common Bean Predict Widespread Divergent Adaptation to Drought. Front. Plant. Sci. 2018, 9, 128.
- Vavilov, N.I. The law of homologous series in variation. J. Genet. 1922, 12, 47–89.
- Tack, J.; Barkley, A.; Nalley, L.L. Effect of warming temperatures on US wheat yields. Proc. Natl. Acad. Sci. USA 2015, 112, 6931–6936.
- Beebe, S.; Rao, I.M.; Cajiao, C.; Grajales, M. Selection for Drought Resistance in Common Bean Also Improves Yield in Phosphorus Limited and Favorable Environments. Crop. Sci. 2008, 48, 582–592.
- Buitrago-Bitar, M.A.; Cortés, A.J.; López-Hernández, F.; Londoño-Caicedo, J.M.; Muñoz-Florez, J.E.; Muñoz, L.C.; Blair, M.W. Allelic Diversity at Abiotic Stress Responsive Genes in Relationship to Ecological Drought Indices for Cultivated Tepary Bean, Phaseolus acutifolius A. Gray, and Its Wild Relatives. Genes 2021, 12, 556.
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005, 24, 23–58.
- Grene, R.; Provart, N.J.; Pardo, J.M. Editorial: Resistance to Salinity and Water Scarcity in Higher Plants. Insights From Extremophiles and Stress-Adapted Plants: Tools, Discoveries and Future Prospects. Front. Plant Sci. 2019, 10, 373.
- Bechtold, U. Plant Life in Extreme Environments: How Do You Improve Drought Tolerance? Front. Plant Sci. 2018, 9, 543.
- Blair, M.W.; Cortés, A.J.; This, D. Identification of an ERECTA gene and its drought adaptation associations with wild and cultivated common bean. Plant. Sci. 2016, 242, 250–259.
- Cortés, A.J.; Chavarro, M.C.; Madriñán, S.; This, D.; Blair, M.W. Molecular ecology and selection in the drought-related Asr gene polymorphisms in wild and cultivated common bean (Phaseolus vulgaris L.). BMC Genet. 2012, 13.
- Cortés, A.J.; This, D.; Chavarro, C.; Madriñán, S.; Blair, M.W. Nucleotide diversity patterns at the drought-related DREB2 encoding genes in wild and cultivated common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2012, 125, 1069–1085.
- Asfaw, A.; Ambachew, D.; Shah, T.; Blair, M.W. Trait Associations in Diversity Panels of the Two Common Bean (Phaseolus vulgaris L.) Gene Pools Grown under Well-watered and Water-Stress Conditions. Front. Plant Sci. 2017, 8, 733.
- Darwin, C. The Variation of Animals and Plants under Domestication; John Murray: London, UK, 1868.
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848.
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852.
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48.
- Khoury, C.K.; Achicanoy, H.A.; Bjorkman, A.D.; Navarro-Racines, C.; Guarino, L.; Flores-Palacios, X.; Engels, J.M.M.; Wiersema, J.H.; Dempewolf, H.; Sotelo, S.; et al. Origins of food crops connect countries worldwide. Proc. R. Soc. B 2016, 283, 1832.
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713.
- Cortés, A.J.; Skeen, P.; Blair, M.W.; Chacón-Sánchez, M.I. Does the genomic landscape of species divergence in Phaseolus beans coerce parallel signatures of adaptation and domestication? Front. Plant Sci. 2018, 9, 1816.
- Stern, D.L.; Orgogozo, V. Is Genetic Evolution Predictable? Science 2009, 323, 746–751.
- Mather, K.A.; Molina, J.; Flowers, J.M.; Rubinstein, S.; Rauh, B.L.; Lawton-Rauh, A.M.Y.; Caicedo, A.L.; McNally, K.L.; Purugganan, M.D. Migration, isolation and hybridization in island crop populations: The case of Madagascar rice. Mol. Ecol. 2010, 19, 4892–4905.
- Marques, D.A.; Meier, J.I.; Seehausen, O. A Combinatorial View on Speciation and Adaptive Radiation. Trends Ecol. Evol. 2019, 34, 531–544.
- Seehausen, O.; Butlin, R.K.; Keller, I.; Wagner, C.E.; Boughman, J.W.; Hohenlohe, P.A.; Peichel, C.L.; Saetre, G.-P.; Bank, C.; Brännström, Å.; et al. Genomics and the origin of species. Nat. Rev. Genet. 2014, 15, 176–192.
- Wolf, J.B.; Ellegren, H. Making sense of genomic islands of differentiation in light of speciation. Nat. Rev. Genet. 2017, 18, 87–100.
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant. Sci. 2010, 15, 684–692.
- Denison, R.F. Darwinian Agriculture: How Understanding Evolution Can Improve Agriculture; Princeton University Press: Princeton, NJ, USA, 2016.
- Tanksley, S.D.; McCouch, S.R. Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild. Science 1997, 227, 1063–1066.
- Ramirez-Villegas, J.; Khoury, C.K.; Achicanoy, H.A.; Mendez, A.C.; Diaz, M.V.; Sosa, C.C.; Debouck, D.G.; Kehel, Z.; Guarino, L. A gap analysis modelling framework to prioritize collecting for ex situ conservation of crop landraces. Divers. Distrib. 2020, 26, 730–742.
- Castaneda-Alvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Maxted, N.; et al. Global conservation priorities for crop wild relatives. Nat. Plants 2016, 2, 16022.
- Salcedo-Castaño, J.; Araya-Villalobos, R.; Castañeda-Alvarez, N.; Toro-Chica, O.; Debouck, D.G. Phaseolus hygrophilus (Leguminosae-Papilionoideae), a new wild bean species from the wet forests of Costa Rica, with notes about section Brevilegumeni. J. Bot. Res. Inst. Tex. 2011, 5, 53–65.
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Fonseca, G.A.B.d.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858.
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.M.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science 2014, 344, 6187.
- Berg, J. Gene-environment interplay. Science 2016, 354, 15.
- Franks, S.J.; Hoffmann, A.A. Genetics of Climate Change Adaptation. Annu. Rev. Genet. 2012, 46, 185–208.
- Stapley, J.; Reger, J.; Feulner, P.G.D.; Smadja, C.; Galindo, J.; Ekblom, R.; Bennison, C.; Ball, A.D.; Beckerman, A.P.; Slate, J. Adaptation genomics: The next generation. Trends Ecol. Evol. 2011, 25, 705–712.
- Hancock, A.M.; Brachi, B.; Faure, N.; Horton, M.W.; Jarymowycz, L.B.; Sperone, F.G.; Toomajian, C.; Roux, F.; Bergelson, J. Adaptation to Climate Across the Arabidopsis thaliana Genome. Science 2011, 334, 83–86.
- Lasky, J.R.; Des Marais, D.L.; McKay, J.K.; Richards, J.H.; Juenger, T.E.; Keitt, T.H. Characterizing genomic variation of Arabidopsis thaliana: The roles of geography and climate. Mol. Ecol. 2012, 21, 5512–5529.
- Cortés, A.J.; Monserrate, F.; Ramírez-Villegas, J.; Madriñán, S.; Blair, M.W. Drought Tolerance in Wild Plant Populations: The Case of Common Beans (Phaseolus vulgaris L.). PLoS ONE 2013, 8, e62898.
- López-Hernández, F.; Cortés, A.J. Last-Generation Genome–Environment Associations Reveal the Genetic Basis of Heat Tolerance in Common Bean (Phaseolus vulgaris L.). Front. Genet. 2019, 10, 22.
- Yoder, J.B.; Stanton-Geddes, J.; Zhou, P.; Briskine, R.; Young, N.D.; Tiffin, P. Genomic signature of adaptation to climate in Medicago truncatula. Genetics 2014, 196, 1263–1275.
- Lasky, J.R.; Upadhyaya, H.D.; Ramu, P.; Deshpande, S.; Hash, C.T.; Bonnette, J.; Juenger, T.E.; Hyma, K.; Acharya, C.; Mitchell, S.E.; et al. Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 2015, 1, e1400218.
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918.
- Forester, B.R.; Jones, M.R.; Joost, S.; Landguth, E.L.; Lasky, J.R. Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 2016, 25, 104–120.
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015, 24, 4348–4370.
- Zahn, L.M.; Purnell, B.A. Genes under pressure. Science 2016, 354, 52.
- Antao, T.; Lopes, A.; Lopes, R.J.; Beja-Pereira, A.; Luikart, G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinform. 2008, 9, 323.
- Kruglyak, L. The road to genome-wide association studies. Nat. Rev. Genet. 2008, 9, 314–318.
- Pennisi, E. Disputed islands. Science 2014, 345, 611–613.
- Maher, B. The case of the Missing Heritability. Nature 2008, 456, 18–21.
- Lambert, C.G.; Black, L.J. Learning from our GWAS mistakes: From experimental design to scientific method. Biostatistics 2012, 13, 195–203.
- Abebe, T.D.; Naz, A.A.; Leon, J. Landscape genomics reveal signatures of local adaptation in barley (Hordeum vulgare L.). Front. Plant Sci. 2015, 6, 813.
- Pluess, A.R.; Frank, A.; Heiri, C.; Lalague, H.; Vendramin, G.G.; Oddou-Muratorio, S. Genome-environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica. New Phytol. 2016, 210, 589–601.
- Frachon, L.; Bartoli, C.; Carrere, S.; Bouchez, O.; Chaubet, A.; Gautier, M.; Roby, D.; Roux, F. A Genomic Map of Climate Adaptation in Arabidopsis thaliana at a Micro-Geographic Scale. Front. Plant Sci. 2018, 9, 967.
- Cortés, A.J.; Wheeler, J.A.; Sedlacek, J.; Lexer, C.; Karrenberg, S. Genome-wide patterns of microhabitat-driven divergence in the alpine dwarf shrub Salix herbacea L. In On The Big Challenges of a Small Shrub: Ecological Genetics of Salix herbacea L.; Acta Universitatis Upsaliensis: Uppsala, Sweden, 2015.
- Muñoz, L.C.; Blair, M.W.; Duque, M.C.; Tohme, J.; Roca, W. Introgression in common bean x tepary bean interspecific congruity-backcross lines as measured by AFLP markers. Crop. Sci. 2003, 44, 637–645.
More
