Hidradenitis suppurativa is a chronic disease of the pilosebaceous unit. The name of the condition is a testament to the presumed relationship between the disease and the microbiome. The pathophysiology of hidradenitis suppurativa is, however, complex and believed to be the product of a multifactorial interplay between the interfollicular epithelium, pilosebaceous unit, microbiome, as well as genetic and environmental factors.
- hidradenitis suppurativa
- microbiome
1. Introduction
Hidradenitis Suppurativa (HS) is a complex condition of the pilosebaceous unit (PSU). Patients experience multiple nodules, abscesses, and draining fistulae in intertriginous skin, which cause a significant impact on their quality of life [1]. The pathophysiology of HS is multifaceted and is the product of interactions between inflammation, a genetic tendency, the microbiome, and environmental factors [2]. The name Hidradenitis Suppurativa is misleading as it suggests the condition to be the result of purely infective processes involving the apocrine glands. This name is the subject of controversy, with some recommending that the disease should be renamed Acne Inversa [3,4][3][4]. The latter name conceptualizes the fact that unlike acne, patients with HS experience deep seated abscesses which involve skin that is not usually affected by acne (intertriginous skin), and which therefore hosts a distinct microbiome [5]. The propensity of HS to manifest as a purulent disease involving apocrine gland-bearing skin is undeniable. However, dismissing the pathophysiological processes at play as purely infective would be an oversimplification.
The intricate role of the microbiome has been investigated in various cutaneous disorders, including psoriasis, atopic dermatitis, and other diseases of the PSU such as acne [6,7,8][6][7][8]. In this review, we scrutinize the existing literature with regard to how the skin microbiome in HS skin differs from that in healthy skin, whether it incites or potentiates disease, how it alters in response to therapy, and whether it can be manipulated to alter disease trajectory.
2. Analysis on Research Results
As the body’s largest interface with the external environment, the skin is home to an extensive array of symbiotic and commensal microorganisms. The skin microbiota orchestrates several key homeostatic processes, including protection against pathogens, immune development, and the breakdown of natural products through a bidirectional metabolic exchange with host cells [36,37][9][10]. Disruption of the natural balance between commensal and pathogenic microbes can lead to skin or even systemic disease, and thus a detailed understanding of the properties of healthy skin microbiome is presented next.
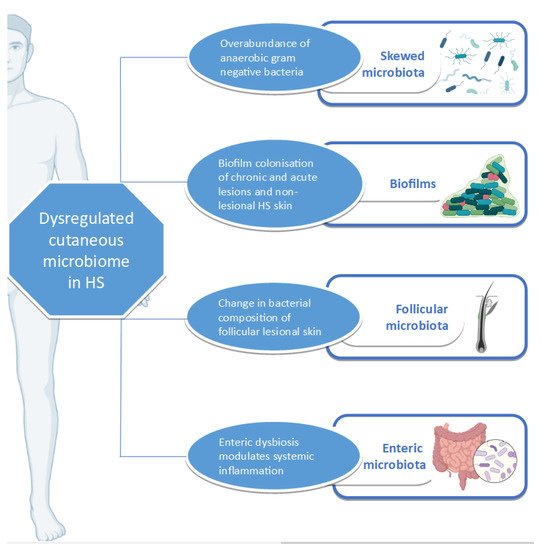
HS skin appears to have a microbiome distinct from that of healthy skin and which is overwhelmingly populated by anaerobic Gram-negative bacteria [68][11]. Differences in HS microbiome can be detected even in the absence of clinically observable lesions [69][12]. Differential abundance gene testing (which refers to the statistical method of investigating the significance of specific bacterial species abundance in two different ecosystems) showed significant differences between lesional and healthy (control) skin as well as between lesional and non-lesional skin [70][13]. The comparison of non-lesional and healthy skin, however, shows no significant difference in bacterial species abundance [71][14]. Functional analysis also confirms significant metagenomic differences between lesional and healthy control skin as well as between lesional and non-lesional skin, but not between non-lesional and healthy control skin [71][14]. The microbiome can vary even between HS-lesion type, with Staphylococcuslugdunesis being associated with nodules and abscesses, polymicrobial anaerobes with chronic suppurating lesions, and anaerobic Actinomycetes with severe suppurating lesions [72][15].
Certain processes favour the tendency of HS skin to host a biofilm. S. epidermidis , a usually non-virulent commensal in healthy skin, has been shown to have a propensity for planktonic growth and biofilm production in both lesional and non-lesional HS skin [90][16]. This bacterium is the abundant CNS in HS patients [91][17]. A matter of concern is S. epidermis in lesional skin, which demonstrates resistance to guideline-recommended antibiotics such as tetracyclines and clindamycin [90,92][16][18]. The biofilm-producing potential of other CNS such as Staphylococcus lugdunensis is also enhanced in HS-lesional skin when compared to non-lesional, control, and reference strains [93][19].
Although HS manifests in the skin, other microbiomes such as the gut microbiome have the potential of mediating disease by facilitating systemic inflammatory pathways supported by the gut–skin axis [101][20]. This pathophysiological axis has been characterized in atopic dermatitis, psoriasis, and rosacea [102,103,104][21][22][23]. Enteric microbiota can influence the skin microbiota, resulting in cutaneous dyshomeostasis, which can ultimately lead to skin pathology, including that of the PSU such as acne [101][20].
3. Current Insights
A complex relationship exists between the microbiome, inflammation, and HS ( Figure 1 ). HS, as an inflammatory condition, is subject to the effects of dysbiosis, whether cutaneous or enteric. Various studies have investigated the relationship between the disease and the microbiome, using an array of sampling and detection methods. Apart from highlighting the yearning of the HS research community to fully understand the pathophysiology of HS, they also bring to the fore the importance of global harmonization in sampling methodology in HS research [137][24].
Beyond its role in pathophysiology, the microbiome can potentially be targeted as a treatment option for altering disease trajectory. It has been shown that most antibiotics used in the treatment of HS exert anti-inflammatory properties. However, research is lacking with regard to the alteration of the microbiome after antibiotic therapy or any therapeutic intervention [138][25]. The use of dietary modification as a therapeutic intervention in the management of disease has several advantages, including the lack of associated negative effects and a low financial burden on the patient. The care of patients with restrictive diets should be overseen by nutritionists, who are ideally familiar with HS and its management as part of the multidisciplinary team providing personalized care to HS patients. As outlined above, advances in genomic technologies can facilitate the bench-to-bedside transition by guiding the management of complex HS cases. Apart from its therapeutic roles, the microbiome can potentially serve as a disease biomarker for the facilitation of disease classification, patient stratification, and the monitoring of treatment response. However, further research is required for the identification and validation of such tools. Furthermore, the microbiome can also help refine genotype-phenotype associations, this being another lacuna in our understanding of HS [139][26].
The studies outlined in this review provide compelling evidence for the dysregulation of the lesional and peri-lesional skin microbiome in HS. However, a direct cause–effect relationship cannot be assumed, and several questions arise. Primarily, the role of non-lesional cutaneous microbiota in the pathogenesis of HS is not completely understood. More specifically, the characterization of the evolution of microbial composition and abundance during the transition from unaffected to lesional skin should provide insight into causative trajectories leading to HS. One can hypothesize on whether changes in microbiota antedate the development of HS, thus representing a latent or pre-clinical stage of the disease. Alternatively, it is possible that the observed cutaneous dysbiosis in HS partly reflects an adaptive microbial response to acute inflammation in active HS lesions. Physiologically, it is likely that the microbiota contributes to causality, progression, and the maintenance of HS against a complex background of host immune response. These factors jointly determine the ensuing clinical picture and response to therapy.
In conclusion, HS is a complex condition, with changes preceding clinical signs and symptoms, and differing according to lesion and anatomical location. Possible future research can be directed towards investigating whether environmental exposure (the hygiene hypothesis) in childhood is related to dysbiosis and disease trajectory in later life. An elegant unifying pathophysiological explanation for HS has not yet been established. Unravelling the role of the microbiome in HS will bring us a step closer to this end.
References
- Matusiak, Ł.; Bieniek, A.; Szepietowski, J.C. Hidradenitis suppurativa markedly decreases quality of life and professional activity. J. Am. Acad. Dermatol. 2010, 62, 706–708.
- Zouboulis, C.C.; Benhadou, F.; Byrd, A.S.; Chandran, N.S.; Giamarellos-Bourboulis, E.J.; Fabbrocini, G.; Frew, J.W.; Fujita, H.; González-López, M.A.; Guillem, P.; et al. What causes hidradenitis suppurativa?—15 years after. Exp. Dermatol. 2020, 29, 1154–1170.
- Sellheyer, K.; Krahl, D. “Hidradenitis suppurativa” is acne inversa! An appeal to (finally) abandon a misnomer. Int. J. Dermatol. 2005, 44, 535–540.
- Scheinfeld, N. Hidradenitis should not be renamed acne inversa. Dermatol. Online J. 2006, 12, 6.
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155.
- Dréno, B.; Dagnelie, M.A.; Khammari, A.; Corvec, S. The Skin Microbiome: A New Actor in Inflammatory Acne. Am. J. Clin. Dermatol. 2020, 21, 18–24.
- Benhadou, F.; Mintoff, D.; Schnebert, B.; Thio, H.B. Psoriasis and Microbiota: A Systematic Review. Diseases 2018, 6, 47.
- Pothmann, A.; Illing, T.; Wiegand, C.; Hartmann, A.A.; Elsner, P. The Microbiome and Atopic Dermatitis: A Review. Am. J. Clin. Dermatol. 2019, 20, 749–761.
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959.
- Gribbon, E.M.; Cunliffe, W.J.; Holland, K.T. Interaction of Propionibacterium acnes with skin lipids in vitro. J. Gen. Microbiol. 1993, 139, 1745–1751.
- Ring, H.C.; Thorsen, J.; Fuursted, K.; Bjarnsholt, T.; Bay, L.; Saunte, D.M.; Thomsen, S.F.; Jemec, G.B. Probiotics in Hidradenitis Suppurativa: A potential treatment option? Clin. Exp. Dermatol. 2021.
- Naik, H.B.; Jo, J.-H.; Paul, M.; Kong, H.H. Skin Microbiota Perturbations Are Distinct and Disease Severity-Dependent in Hidradenitis Suppurativa. J. Invest. Dermatol. 2020, 140, 922–925.
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 1–18.
- Ring, H.C.; Thorsen, J.; Jørgensen, A.H.; Bay, L.; Bjarnsholt, T.; Fuursted, K.; Thomsen, S.F.; Jemec, G.B. Predictive Metagenomic Analysis Reveals a Role of Cutaneous Dysbiosis in the Development of Hidradenitis Suppurativa. J. Invest. Dermatol. 2020, 140, 1473–1476.
- Guet-Revillet, H.; Coignard-Biehler, H.; Jais, J.-P.; Quesne, G.; Frapy, E.; Poirée, S.; Guern, A.-S.L.; Flèche-Matéos, A.L.; Hovnanian, A.; Consigny, P.-H.; et al. Bacterial pathogens associated with hidradenitis suppurativa, France. Emerg. Infect. Dis. 2014, 20, 1990–1998.
- Ardon, C.B.; Prens, E.P.; Fuursted, K.; Ejaz, R.N.; Shailes, J.; Jenssen, H.; Jemec, G.B.E. Biofilm production and antibiotic susceptibility of Staphylococcus epidermidis strains from Hidradenitis Suppurativa lesions. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 170–177.
- Nikolakis, G.; Liakou, A.I.; Bonovas, S.; Seltmann, H.; Bonitsis, N.; Join-Lambert, O.; Wild, T.; Karagiannidis, I.; Zolke-Fischer, S.; Langner, K.; et al. Bacterial Colonization in Hidradenitis Suppurativa/Acne Inversa: A Cross-sectional Study of 50 Patients and Review of the Literature. Acta Derm. Venereol. 2017, 97, 493–498.
- Hendricks, A.J.; Hsiao, J.L.; Lowes, M.A.; Shi, V.Y. A Comparison of International Management Guidelines for Hidradenitis Suppurativa. Dermatology 2021, 237, 81–96.
- Ardon, C.B.; Prens, E.P.; Tkadlec, J.; Fuursted, K.; Abourayale, S.; Jemec, G.B.E.; Jenssen, H. Virulent Staphylococcus lugdunensis with limited genetic diversity in hidradenitis suppurativa lesions. J. Eur. Acad. Dermatol. Venereol. JEADV 2019, 33, e248–e250.
- Salem, I.; Ramser, A.; Isham, N.; Ghannoum, M.A. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front. Microbiol. 2018, 9, 1459.
- Lee, S.Y.; Lee, E.; Park, Y.M.; Hong, S.J. Microbiome in the Gut-Skin Axis in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2018, 10, 354–362.
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 3201.
- Searle, T.; Ali, F.R.; Carolides, S.; Al-Niaimi, F. Rosacea and the gastrointestinal system. Australas. J. Dermatol. 2020, 61, 307–311.
- Naik, H.B.; Piguet, V. Standardizing Hidradenitis Suppurativa Skin Microbiome Research: The Methods Matter. J. Invest. Dermatol. 2020, 140, 1688–1690.
- Frew, J.W.; Hawkes, J.E.; Krueger, J.G. Topical, systemic and biologic therapies in hidradenitis suppurativa: Pathogenic insights by examining therapeutic mechanisms. Ther. Adv. Chronic Dis. 2019, 10, 2040622319830646.
- Assan, F.; Bachelez, H. Reply. J. Allergy Clin. Immunol. 2020, 146, 461–462.
