Epigenetic modifications are able to alter gene expression and include DNA methylation, different histone variants, and post-transcriptional modifications (PTMs), such as acetylation or phosphorylation, and through short/long RNAs, respectively. In this review, we focus on current knowledge concerning epigenetic modifications in gene regulation. We describe different forms of epigenetic modifications and explain how epigenetic changes can be detected. The relevance of epigenetics in renal diseases is highlighted with multiple examples and the use of the zebrafish model to study glomerular diseases in general and epigenetics in renal diseases in particular is discussed. We end with an outlook on how to use epigenetic modifications as a therapeutic target for different diseases. Here, the zebrafish model can be employed as a high-throughput screening tool not only to discover epigenetic alterations contributing to disease, but also to test novel substances that change epigenetic signatures in vivo. Therefore, the zebrafish model harbors the opportunity to find novel pathogenic pathways allowing a pre-selection of potential targets and compounds to be tested for renal diseases.
- zebrafish model
- epigenetics
- renal diseases
- microRNAs
1. Epigenetic Modifications
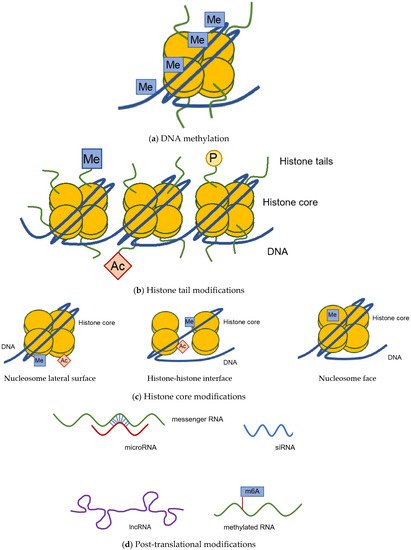
1.1. DNA Methylation
1.2. Histone Modification
1.3. Post-Transcriptional Modification
2. Using the Zebrafish Model to Study Epigenetics in Renal Diseases
In recent years, the zebrafish (Danio rerio) model has been used to study glomerular function and disease [19][20][21][22][23]. The zebrafish larvae’s pronephros, which is composed of two bilateral pronephric ducts linked with fused glomeruli in the midline of the larvae, is very similar to the human metanephros [24][25]. The pronephros tubular epithelium is composed of two proximal convoluted tubules, two proximal straight tubules, two distal early and distal late tubule segments, and a pronephric duct [26]. The main difference between the pronephros of the zebrafish and the mammalian partner is that the pronephros does not have a thin limb segment between the proximal straight tubule and the thick ascending limb. The glomerulus of the pronephros contains podocytes, glomerular basement membrane fenestrated endothelial cells, and mesangial cells [21]. Glomerular filtration begins as early as 48 h post-fertilization (hpf) and a fully functioning pronephros of zebrafish larvae is fully developed within 72 hpf [27][28]. At least 70% of zebrafish proteins have a human orthologue [29]. Furthermore, the zebrafish is very amenable to genetic manipulations though microinjections of morpholinos, DNAs, RNAs, and microRNAs [19][21][22][23][30][31][32][33][34]. Given its high genetic and renal similarity to humans the zebrafish has been used to study different renal diseases such as FSGS [35][36][37][38], polycystic kidney diseases [39][40][41], diabetic nephropathy [38][42], and renal cancer [43]. However, these manipulations can have effects on the developing embryo and observed phenotypes might not be specific to the tissue or organ of interest. Furthermore, injection of these molecules often has off-target effects, which are sometimes difficult to identify. For more specificity, genetic editing techniques such as transcription activator-like effector nucleases (TALENs), zinc finger nucleases, and clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 can also be performed in the zebrafish [44][45][46]. The zebrafish has already served as a model to investigate epigenetic changes in hearing loss [47], development [48], and cancer [43][49][50] and in propagating secondary complications observed in diabetes mellitus [51]. However, though the zebrafish model has been used abundantly in kidney research, so far only few studies have focused on the epigenetic contribution to renal diseases in this versatile in vivo model [19][22][23][52].2.1. DNA Methylation
2.2. Histone Modifications
2.3. Post-Transcriptional Modifications
| Study | Renal Disease | Epigenetic Mechanisms/Transcription Factors Involved | Results | |
|---|---|---|---|---|
| “Overexpression of TGF-β Inducible microRNA-143 in Zebrafish Leads to Impairment of the Glomerular Filtration Barrier by Targeting Proteoglycans”; Müller-Deile et al. | [22] | FSGS (focal segmental glomerulosclerosis) | Downregulation of versian and syndecan by miR-143-3p | Proteinuria, edema, and podocyte effacement |
| “Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases”; Müller-Deile et al. | [19] | Membranous glomerulonephritis | Downregulation of nephronectin by miR-367a-3p | Proteinuria, edema, podocyte effacement, and disrupted glomerular basement membrane |
| “Overexpression of preeclampsia-induced microRNA-26a-5p leads to proteinuria in zebrafish”; Müller-Deile et al. | [23] | Preeclampsia | Downregulation of vascular endothelial growth factor A (VEGF-A) by miR-26a-5p | Proteinuria, edema, and glomerular endotheliosis |
| “Chromatin architecture reveals cell-type-specific target genes for kidney disease risk variants”; Duan et al. | [49] | Risk variants for renal tumor and chronic kidney disease | Histone modifications of risk variants | Renal tumor and chronic kidney disease |
| “Activation of P-TEFb by cAMP-PKA signaling in autosomal dominant polycystic kidney disease”; Sun et al. | [58] | ADPKD (autosomal dominant polycystic kidney disease) | cAMP-PKA signaling disrupts the inactive P-TEFb/HEXIM1/7SK snRNP complex | Cystogenesis |
| “Wolf–Hirschhorn syndrome candidate 1-like 1 epigenetically regulates nephrin gene expression”; Ito et al. | [55] | Nephrotic syndrome | Wolf–-Hirschhorn syndrome candidate 1-like (WHSC1L1-L) acts as a histone methyltransferase and regulates nephrin gene expression | Reduction of nephrin mRNA |
| “Loss of vhl in the zebrafish pronephros recapitulates early stages of human clear cell renal cell carcinoma”; Noonan et al. | [43] | Clear cell renal cell carcinoma | von Hippel-Lindau (vhl) inactivation leads to > Stabilization of hypoxia-inducible factors 1a and 2a (HIF1a and HIF2a) > Upregulation of specific target genes involved in cell proliferation, angiogenesis and erythropoiesis |
Increased tubule diameter, disorganized cilia, cytoplasmic lipid vesicles, glycogen accumulation, aberrant cell proliferation, and abnormal apoptosis |
| “The transcription factor Dach1 is essential for podocyte function”; Endlich et al. | [59] | Podocyte differentiation and proper kidney function | Transcription factor Dach1 | Downregulation of nephrin, edema, and leakage of the filtration barrier |
| “Mutation of microphthalmia-associated transcription factor (mitf) in zebrafish sensitizes for glomerulopathy”; Müller-Deile et al. | [52] | Glomerulopathy | Mutation in microphthalmia-associated transcription factor (mitf) | Increased susceptibility to edema, ptoteinuria, and podocyte effacement after puromycin treatment |
References
- Wu, T.P.; Wang, T.; Seetin, M.G.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.D.; Mackintosh, S.G.; Zhong, M.; et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016, 532, 329–333.
- Mersfelder, E.L.; Parthun, M.R. The tale beyond the tail: Histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006, 34, 2653–2662.
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307.
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200.
- Coco, C.; Sgarra, L.; Potenza, M.A.; Nacci, C.; Pasculli, B.; Barbano, R.; Parrella, P.; Montagnani, M. Can Epigenetics of Endothelial Dysfunction Represent the Key to Precision Medicine in Type 2 Diabetes Mellitus? Int. J. Mol. Sci. 2019, 20, 1949.
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022.
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607.
- Parry, A.; Rulands, S.; Reik, W. Active turnover of DNA methylation during cell fate decisions. Nat. Rev. Genet. 2021, 22, 59–66.
- Kulis, M.; Merkel, A.; Heath, S.; Queiros, A.C.; Schuyler, R.P.; Castellano, G.; Beekman, R.; Raineri, E.; Esteve, A.; Clot, G.; et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat. Genet. 2015, 47, 746–756.
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905.
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322.
- Yun, M.; Wu, J.; Workman, J.L.; Li, B. Readers of histone modifications. Cell Res. 2011, 21, 564–578.
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260.
- Tropberger, P.; Schneider, R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 657–661.
- Brennecke, J.; Malone, C.D.; Aravin, A.A.; Sachidanandam, R.; Stark, A.; Hannon, G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008, 322, 1387–1392.
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009, 457, 405–412.
- Siomi, H.; Siomi, M.C. On the road to reading the RNA-interference code. Nature 2009, 457, 396–404.
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42.
- Müller-Deile, J.; Dannenberg, J.; Schroder, P.; Lin, M.H.; Miner, J.H.; Chen, R.; Bräsen, J.H.; Thum, T.; Nyström, J.; Staggs, L.B.; et al. Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases. Kidney Int. 2017, 92, 836–849.
- Morales, E.E.; Wingert, R.A. Zebrafish as a Model of Kidney Disease. Results Probl. Cell Differ. 2017, 60, 55–75.
- Schenk, H.; Masseli, A.; Schroder, P.; Bolanos-Palmieri, P.; Beese, M.; Hegermann, J.; Brasen, J.H.; Haller, H. Sulfatases, in Particular Sulf1, Are Important for the Integrity of the Glomerular Filtration Barrier in Zebrafish. BioMed Res. Int. 2019, 2019, 4508048.
- Muller-Deile, J.; Gellrich, F.; Schenk, H.; Schroder, P.; Nystrom, J.; Lorenzen, J.; Haller, H.; Schiffer, M. Overexpression of TGF-beta Inducible microRNA-143 in Zebrafish Leads to Impairment of the Glomerular Filtration Barrier by Targeting Proteoglycans. Cell Physiol. Biochem. 2016, 40, 819–830.
- Muller-Deile, J.; Schroder, P.; Beverly-Staggs, L.; Hiss, R.; Fiedler, J.; Nystrom, J.; Thum, T.; Haller, H.; Schiffer, M. Overexpression of preeclampsia induced microRNA-26a-5p leads to proteinuria in zebrafish. Sci. Rep. 2018, 8, 3621.
- Drummond, I.A. The zebrafish pronephros: A genetic system for studies of kidney development. Pediatr. Nephrol. 2000, 14, 428–435.
- Naylor, R.W.; Qubisi, S.S.; Davidson, A.J. Zebrafish Pronephros Development. Results Probl. Cell Differ. 2017, 60, 27–53.
- Wingert, R.A.; Davidson, A.J. The zebrafish pronephros: A model to study nephron segmentation. Kidney Int. 2008, 73, 1120–1127.
- Hanke, N.; Staggs, L.; Schroder, P.; Litteral, J.; Fleig, S.; Kaufeld, J.; Pauli, C.; Haller, H.; Schiffer, M. “Zebrafishing” for novel genes relevant to the glomerular filtration barrier. BioMed Res. Int. 2013, 2013, 658270.
- Drummond, I.A.; Majumdar, A.; Hentschel, H.; Elger, M.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.; Stemple, D.L.; Zwartkruis, F.; Rangini, Z.; et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 1998, 125, 4655–4667.
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503.
- Kawakami, K.; Takeda, H.; Kawakami, N.; Kobayashi, M.; Matsuda, N.; Mishina, M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 2004, 7, 133–144.
- Langheinrich, U.; Hennen, E.; Stott, G.; Vacun, G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr. Biol. 2002, 12, 2023–2028.
- Li, Q.; Sadowski, S.; Frank, M.; Chai, C.; Varadi, A.; Ho, S.Y.; Lou, H.; Dean, M.; Thisse, C.; Thisse, B.; et al. The abcc6a gene expression is required for normal zebrafish development. J. Investig. Dermatol. 2010, 130, 2561–2568.
- Yoruk, B.; Gillers, B.S.; Chi, N.C.; Scott, I.C. Ccm3 functions in a manner distinct from Ccm1 and Ccm2 in a zebrafish model of CCM vascular disease. Dev. Biol. 2012, 362, 121–131.
- Patra, C.; Diehl, F.; Ferrazzi, F.; van Amerongen, M.J.; Novoyatleva, T.; Schaefer, L.; Muhlfeld, C.; Jungblut, B.; Engel, F.B. Nephronectin regulates atrioventricular canal differentiation via Bmp4-Has2 signaling in zebrafish. Development 2011, 138, 4499–4509.
- Hansen, K.U.I.; Siegerist, F.; Daniel, S.; Schindler, M.; Iervolino, A.; Blumenthal, A.; Daniel, C.; Amann, K.; Zhou, W.; Endlich, K.; et al. Prolonged podocyte depletion in larval zebrafish resembles mammalian focal and segmental glomerulosclerosis. FASEB J. 2020, 34, 15961–15974.
- Kotb, A.M.; Simon, O.; Blumenthal, A.; Vogelgesang, S.; Dombrowski, F.; Amann, K.; Zimmermann, U.; Endlich, K.; Endlich, N. Knockdown of ApoL1 in Zebrafish Larvae Affects the Glomerular Filtration Barrier and the Expression of Nephrin. PLoS ONE 2016, 11, e0153768.
- Schiffer, M.; Teng, B.; Gu, C.; Shchedrina, V.A.; Kasaikina, M.; Pham, V.A.; Hanke, N.; Rong, S.; Gueler, F.; Schroder, P.; et al. Pharmacological targeting of actin-dependent dynamin oligomerization ameliorates chronic kidney disease in diverse animal models. Nat. Med. 2015, 21, 601–609.
- Teng, B.; Schroder, P.; Muller-Deile, J.; Schenk, H.; Staggs, L.; Tossidou, I.; Dikic, I.; Haller, H.; Schiffer, M. CIN85 Deficiency Prevents Nephrin Endocytosis and Proteinuria in Diabetes. Diabetes 2016, 65, 3667–3679.
- Kim, E.; Arnould, T.; Sellin, L.K.; Benzing, T.; Fan, M.J.; Gruning, W.; Sokol, S.Y.; Drummond, I.; Walz, G. The polycystic kidney disease 1 gene product modulates Wnt signaling. J. Biol. Chem. 1999, 274, 4947–4953.
- Low, S.H.; Vasanth, S.; Larson, C.H.; Mukherjee, S.; Sharma, N.; Kinter, M.T.; Kane, M.E.; Obara, T.; Weimbs, T. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev. Cell 2006, 10, 57–69.
- Obara, T.; Mangos, S.; Liu, Y.; Zhao, J.; Wiessner, S.; Kramer-Zucker, A.G.; Olale, F.; Schier, A.F.; Drummond, I.A. Polycystin-2 immunolocalization and function in zebrafish. J. Am. Soc. Nephrol. 2006, 17, 2706–2718.
- Sharma, K.R.; Heckler, K.; Stoll, S.J.; Hillebrands, J.L.; Kynast, K.; Herpel, E.; Porubsky, S.; Elger, M.; Hadaschik, B.; Bieback, K.; et al. ELMO1 protects renal structure and ultrafiltration in kidney development and under diabetic conditions. Sci. Rep. 2016, 6, 37172.
- Noonan, H.R.; Metelo, A.M.; Kamei, C.N.; Peterson, R.T.; Drummond, I.A.; Iliopoulos, O. Loss of vhl in the zebrafish pronephros recapitulates early stages of human clear cell renal cell carcinoma. Dis. Model. Mech. 2016, 9, 873–884.
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229.
- Liu, C.X.; Li, C.Y.; Hu, C.C.; Wang, Y.; Lin, J.; Jiang, Y.H.; Li, Q.; Xu, X. CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors. Mol. Autism 2018, 9, 23.
- Moore, F.E.; Reyon, D.; Sander, J.D.; Martinez, S.A.; Blackburn, J.S.; Khayter, C.; Ramirez, C.L.; Joung, J.K.; Langenau, D.M. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). PLoS ONE 2012, 7, e37877.
- Hunt, E.A.; Broyles, D.; Head, T.; Deo, S.K. MicroRNA Detection: Current Technology and Research Strategies. Annu. Rev. Anal. Chem. 2015, 8, 217–237.
- Balasubramanian, S.; Raghunath, A.; Perumal, E. Role of epigenetics in zebrafish development. Gene 2019, 718, 144049.
- Duan, A.; Wang, H.; Zhu, Y.; Wang, Q.; Zhang, J.; Hou, Q.; Xing, Y.; Shi, J.; Hou, J.; Qin, Z.; et al. Chromatin architecture reveals cell type-specific target genes for kidney disease risk variants. BMC Biol. 2021, 19, 38.
- Feitsma, H.; Cuppen, E. Zebrafish as a cancer model. Mol. Cancer Res. 2008, 6, 685–694.
- Sarras, M.P., Jr.; Leontovich, A.A.; Intine, R.V. Use of zebrafish as a model to investigate the role of epigenetics in propagating the secondary complications observed in diabetes mellitus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2015, 178, 3–7.
- Müller-Deile, J.; Dannenberg, J.; Liu, P.; Lorenzen, J.; Nyström, J.; Thum, T.; Schiffer, M. Identification of cell and disease specific microRNAs in glomerular pathologies. J. Cell Mol. Med. 2019, 23, 3927–3939.
- Magnani, E.; Macchi, F.; Madakashira, B.P.; Zhang, C.; Alaydaroos, F.; Sadler, K.C. uhrf1 and dnmt1 Loss Induces an Immune Response in Zebrafish Livers Due to Viral Mimicry by Transposable Elements. Front. Immunol. 2021, 12, 627926.
- Siebenthall, K.T.; Miller, C.P.; Vierstra, J.D.; Mathieu, J.; Tretiakova, M.; Reynolds, A.; Sandstrom, R.; Rynes, E.; Haugen, E.; Johnson, A.; et al. Integrated epigenomic profiling reveals endogenous retrovirus reactivation in renal cell carcinoma. EBioMedicine 2019, 41, 427–442.
- Ito, Y.; Katayama, K.; Nishibori, Y.; Akimoto, Y.; Kudo, A.; Kurayama, R.; Hada, I.; Takahashi, S.; Kimura, T.; Fukutomi, T.; et al. Wolf-Hirschhorn syndrome candidate 1-like 1 epigenetically regulates nephrin gene expression. Am. J. Physiol.-Ren. Physiol. 2017, 312, F1184–F1199.
- Xiao, C.; Wang, F.; Hou, J.; Zhu, X.; Luo, Y.; Xiong, J.W. Nanoparticle-mediated siRNA Gene-silencing in Adult Zebrafish Heart. J. Vis. Exp. 2018, e58054.
- Wu, H.; Zhao, M.; Yoshimura, A.; Chang, C.; Lu, Q. Critical Link Between Epigenetics and Transcription Factors in the Induction of Autoimmunity: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 50, 333–344.
- Sun, Y.; Liu, Z.; Cao, X.; Lu, Y.; Mi, Z.; He, C.; Liu, J.; Zheng, Z.; Li, M.J.; Li, T.; et al. Activation of P-TEFb by cAMP-PKA signaling in autosomal dominant polycystic kidney disease. Sci. Adv. 2019, 5, eaaw3593.
- Endlich, N.; Kliewe, F.; Kindt, F.; Schmidt, K.; Kotb, A.M.; Artelt, N.; Lindenmeyer, M.T.; Cohen, C.D.; Döring, F.; Kuss, A.W.; et al. The transcription factor Dach1 is essential for podocyte function. J. Cell Mol. Med. 2018, 22, 2656–2669.
