Out of the various text mining tasks and techniques, our goal in this paper is to review the current state-of-the-art in Clinical Named Entity Recognition (NER) and Relationship Extraction (RE)-based techniques. Clinical NER is a natural language processing (NLP) method used for extracting important medical concepts and events i.e., clinical NEs from the data. Relationship Extraction (RE) is used for detecting and classifying the annotated semantic relationships between the recognized entities.
- electronic health records
- clinical text
- natural language processing
- named entity recognition
- relationship extraction
- machine learning
1. Introduction
The amount of text generated every day is increasing drastically in different domains such as health care, news articles, scientific literature, and social media. Since 2010, the International Data Corporation (IDC) has predicted that the amount of data can potentially grow 50-fold to 40 billion terabytes by 2020 [1]. Textual data is very common in most domains, but automated comprehension is difficult due to its unstructured nature and has led to the design of several text mining (TM) techniques in the last decade.
TM refers to the extraction of interesting and nontrivial patterns or knowledge from text [2]. Common text mining tasks include text preprocessing, text classification, question-answering, clustering, and statistical techniques.
TM has become extremely popular and useful in the biomedical and healthcare domains. In healthcare, about 80% of the total medical data is unstructured and untapped after its creation [3]. This unstructured data from hospitals, healthcare clinics, or biomedical labs can come in many forms such as text, images, and signals. Out of the various text mining tasks and techniques, our goal in this paper is to review the current state-of-the-art in Clinical Named Entity Recognition (NER) and Relationship Extraction (RE)-based techniques. Clinical NER is a natural language processing (NLP) method used for extracting important medical concepts and events i.e., clinical NEs from the data [4]. Relationship Extraction (RE) is used for detecting and classifying the annotated semantic relationships between the recognized entities. Significant research on NER and RE has been carried out in the past both on clinical narratives and other types of text. For example, in the sentence, “ Her white count remained elevated despite discontinuing her G-CSF ”, the words in bold are the various entities in the sentence. After the entities are recognized, the relationship between two or more entities is extracted. In this case, “ her white count ” and “ elevated ” are found to be related to each other in a manner dissimilar to the nature of the relationship between “ elevated ” and “ her G-CSF ”. In the sentence “ Atorvastatin is found to have therapeutic effects in breast cancer although no clinical trials are performed at present”, the NE of interest includes the name of the drug (atorvastatin) and the disease name (breast cancer), whereas the drug–disease relation (atorvastatin–breast cancer) is the relationship of interest. Figure 1 shows a pictorial representation of the association between NER and RE.
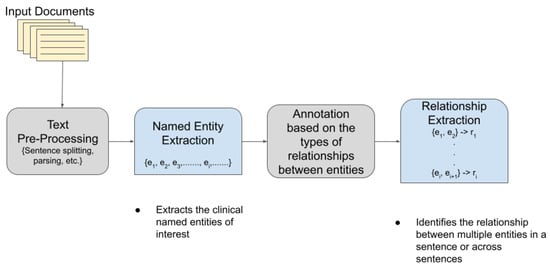
2. NLP Competitions and Datasets for Clinical Text
In this section, we review the different NLP competitions and datasets that are more geared towards clinical text.
The datasets are important in understanding the different entities and relations extracted in the clinical domain. This subsection gives an overview of the different datasets used for clinical NER and RE tasks for a better understanding of the challenges in the domain.
We came across a few publicly available datasets for clinical NER; however, these datasets are restricted to specific NLP tasks in clinical domain. Below is a list of datasets that were used in NER challenges or used as training for NER models, which are discussed in Section 5.3 for training, testing, and validation: Mayo Clinic EMR: It has around 273 clinical notes, which includes 61 consult, 4 educational visits and general medical examinations, and a couple of exam notes. A few models, such as Savova et al. [46][5], generated a clinical corpus from Mayo Clinic EMR [47][6]. MADE1.0 Data set: This dataset consists of 1092 medical notes from 21 randomly selected cancer patients’ EHR notes at the University of Massachusetts Memorial Hospital. FoodBase Corpus: It consists of 1000 recipes annotated with food concepts. The recipes were collected from a popular recipe sharing social network. This is the first annotated corpora with food entities and was used by Popovski et al. [48][7] to compare food-based NER methods and to extract food entities from dietary records for individuals that were written in an unstructured text format. Swedish and Spanish Clinical Corpora [49][8]: This dataset consists of annotated corpora clinical texts extracted from EHRs; the Spanish dataset consists of annotated entities for disease and drugs, while the Swedish dataset has entities annotated for body parts, disorder, and findings. This dataset is mostly used for training and validation for NER on Swedish and Spanish clinical text. i2b2 2010 dataset [38][9]: This dataset includes discharge data summaries from Partners Healthcare, Beth Israel Deaconess Medical center, and University of Pittsburgh (also contributed progress reports). It consists of 394 training, 477 test, and 877 unannotated reports. All of the information are de-identified and released for challenge. These datasets are used for training and validation in many of the NER models used for clinical text. MIMIC-III Clinical Database [50][10] : This is a large and freely available dataset consisting of de-identified clinical data of more than 40,000 patients who stayed at the Beth Israel Deaconess Medical Center between 2001 and 2012. This dataset also consists of free-text notes, besides also providing a demo dataset with information for 100 patients. Shared Annotated Resources (shARe) Corpus [37][11]: This dataset consists of a corpus annotated with disease/disorder in clinical text. CanTeMiST [51][12]: It comprises 6933 clinical documents that does not contain any PHI. The dataset is annotated for the synonyms of tumor morphology and was used for clinical NER on a Spanish text by Vunkili et al. [51][12].
Specific relations annotated in the datasets from the various clinical RE challenges mentioned in Section 4.1 are as follows: 1. 2010 i2b2/VA Challenge on Concepts, Assertions, and Relations in Clinical Text [38][9]: A wide variety of relations were identified as follows: Medical problem–treatment relations: - TrIP indicates that treatment improves medical problems, such as hypertension being controlled by hydrochlorothiazide . - TrWP indicates that treatment worsens medical conditions, such as the tumor growing despite the available chemotherapeutic regimen . - TrCP indicates that treatment causes medical problems, such as Bactrium possibly being a cause of abnormalities . - TrAP indicates that treatment is administered for medical problems, e.g., periodic Lasix treatment preventing congestive heart failure . - TrNAP indicates that treatment is not administered because of medical problems e.g., Relafen being contraindicated because of ulcers . - Others that do not fit into medical problem–treatment relations. Medical problem–test relations: - TeRP indicates that the test reveals medical problems, such as an MRI revealing a C5-6 disc herniation . - TeCP indicates that the test was conducted to investigate a medical problem, such as a VQ scan being performed to investigate a pulmonary embolus . - Others that do not fit into medical–test relations. Medical problem–medical problem relations: - PIP indicates any kind of medical problem such as a C5–6 disc herniation with cord compression . - Other relations with respect to medical problems that do not fit into the PIP relationship. 2. 2011 Evaluating the state-of-the-art in coreference resolution for electronic medical records [40][13]: The data for this challenge was similar to the 2010 i2b2/VA challenge as the dataset contained two separate corpora, i.e., the i2b2/VA corpus and the Ontology Development and Information Extraction (ODIE) corpus, which contained de-identified clinical reports, pathology reports, etc. 3. Evaluating temporal relations in clinical text, 2012 i2b2 Challenge [43][14]: The temporal relations or links in the dataset indicate how two events or two time expressions or an event and a time expression is related to each other. The possible links annotated in the dataset were BEFORE, AFTER, SIMULTANEOUS, OVERLAP, BEGUN_BY, ENDED_BY, DURING, and BEFORE_OVERLAP. Ex: OVERLAP -> She denies any fever or chills . Ex: ENDED_BY -> His nasogastric tube was discontinued on 05-26-98 . 4. 2018 n2c2 shared a task on adverse drug events and medication extraction in electronic health records [44][15]: The different relations identified between two entities in this case are either of the following types: Strength–Drug, Form–Drug, Dosage–Drug, Frequency–Drug, Route–Drug, Duration–Drug, Reason–Drug, and ADE–Drug.
3. Trends and Future Research Directions
Our main observation from this review is that the clinical-NER community is more focused on deep learning as it has shown promising results. The other approaches such as dictionary or rule-based methods have lost popularity in the last few years. We believe that the upcoming research on clinical NER will develop models using hybrid approaches where the ML-based and rule/dictionary-based approaches can be combined. One of the major challenges while evaluating different clinical NER models was how to measure their effectiveness. The F1-score measure has its own limitations, as mentioned earlier; simply comparing the F1-score does not give much insight into the models. We have seen recently that there are few attempts to address the limitations of F1-score and suggest alternative metrics such as [103][16]. However, currently, we did not see any attempts to standardize an evaluation metric for clinical NER. For the class imbalance problem discussed in this survey paper, we believe that the community should consider using metrics that address the multi-class imbalance problem. We did see multiple metrics available; however, the selection of correct metric is based on the user interest towards majority or minority classes. Alternatively, we recommend using multiple metrics to obtain a better idea of the balanced performance. We have seen many recent works published on performing clinical NER on text from different languages apart from English and Chinese text such as [52][17] in Italian text. There are attempts to use transfer learning from the text in different languages to improve the performance such as [52][17]; although this is still in an initial phase, we believe that, in the next few years, more work will follow this approach. As mentioned in the Clinical NER section, one of the major issues in clinical NER is that most of the models developed are only limited to specific clinics or centers, and specific domains. In order to address this and to make clinical NER models widely available for usage, the clinical terms should be standardized and widely adopted. We found a few attempts on the standardization of clinical terms such as [53][18]; however, there is not much work currently available that attempts to perform clinical NER on standardized clinical terms and is available for adoption. We believe that the community will move towards a standardization of clinical terms and that future models developed will aim to use those terms. We also noticed that the clinical NER tasks performed vary based on different domains; our survey found that none of them have used transfer learning approaches to train their models from different domains. We believe that, with the success of transfer learning in [52][17], the community will be looking to develop their deep learning models using transfer learning from different clinical NER tasks.
Most of the clinical NER tasks that we came across aimed to identify the entities from clinical text and then to use them for other NLP tasks. Given the sensitive nature of the clinical text, it is becoming difficult to publish models that are developed for clinical NER. The community is trying to overcome this by developing clinical NER models that identify sensitive terms/entities from clinical text, remove them, and make them available for publishing. Recently, other ML communities are using GANs (Generative Adversarial Networks) [104][19], which automatically discover patterns in the data and can develop synthetic data that looks similar to the actual data. This approach has many benefits such as handling privacy as no real data is compromised or used in a training phase, and it is capable of handling under sampling and oversampling for multiple classes. We believe that, in the future, clinical NER models will use GANs to develop more robust and scalable models. Likewise, this approach can be one of the potential approaches for clinical RE.
NER reconciliation is a process of collecting data from multiple sources, gathering and mapping them to a real-world object. In clinical NER, this problem can be more severe, as in the radiation oncology domain, different physicians can assign different names to the same structure. Most of the datasets discussed in this paper are annotated and follow the standard naming convention, but this process is not scalable if multiple data sources are used for integration. We performed an extensive search to find any literature on clinical NER reconciliation. To date, we did not find any attempts to perform clinical NER reconciliation. However, we found a few attempts for NER reconciliation in other domains such as Isaac et al. [105][20] and Van Holland et al. [106][21]; these approaches are geared towards vocabulary reconciliation. We believe that clinical NER reconciliation is an open research problem. As mentioned earlier, there are ongoing attempts to standardize the clinical terms, and if such a standardization is widely adopted by physicians, it can make the integration process a lot simpler.
After surveying the clinical RE papers, it was found that, lately, the community is most interested in investigating traditional ML-based approaches, deep learning-based approaches, and language models to perform clinical RE. Very little research using rule-based approaches are coming up but unsupervised learning-based methods for clinical RE have become somewhat dormant because of the uncertainty in the results generated by these methods. Rule-based methods were used in many research works before 2016. With the introduction of newer techniques and newer research over the years, the performance of the clinical RE tasks kept on improving. Later on, traditional ML-based methods and deep learning methods along with different feature representation techniques were adopted for this purpose. It was observed that the traditional methods outperformed the deep learning methods in many cases. In some cases, deep learning methods performed poorer than rule-based methods. This may be due to the limited data used in most of these works. Deep learning methods generally perform better than traditional methods in case of a large amount of data, but clinical data is often limited. This is a practical limitation of using deep learning methods for clinical RE. In this era of supervised learning on clinical texts, it was found that the language models such as BERT and its variations vastly perform the best in extracting relations from clinical texts. This shows that the language models are somewhat capable of understanding the intricacies of the language better. However, experimentation with newer and advanced supervised algorithms for relationship classification in the clinical domain should continue in the future as the performance of the algorithms often vary with the data.
References
- Gantz, J.; Reinsel, D. The digital universe in 2020: Big data, bigger digital shadows, and biggest growth in the far east. IDC IView IDC Anal. Future 2012, 2007, 1–16.
- Tan, A.H. Text mining: The state of the art and the challenges. In Proceedings of the Pakdd 1999 Workshop on Knowledge Disocovery from Advanced Databases, Bejing, China, 26–28 April 1999; Volume 8, pp. 65–70.
- Kong, H.J. Managing unstructured big data in healthcare system. Healthc. Inform. Res. 2019, 25, 1–2.
- Wu, Y.; Jiang, M.; Xu, J.; Zhi, D.; Xu, H. Clinical Named Entity Recognition Using Deep Learning Models. AMIA Annu. Symp. Proc. 2017, 2017, 1812–1819.
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513.
- Olson, J.E.; Ryu, E.; Johnson, K.J.; Koenig, B.A.; Maschke, K.J.; Morrisette, J.A.; Liebow, M.; Takahashi, P.Y.; Fredericksen, Z.S.; Sharma, R.G.; et al. The Mayo Clinic Biobank: A building block for individualized medicine. Mayo Clin. Proc. 2013, 88, 952–962.
- Popovski, G.; Seljak, B.K.; Eftimov, T. A survey of named-entity recognition methods for food information extraction. IEEE Access 2020, 8, 31586–31594.
- Weegar, R.; Pérez, A.; Casillas, A.; Oronoz, M. Recent advances in Swedish and Spanish medical entity recognition in clinical texts using deep neural approaches. BMC Med. Inform. Decis. Mak. 2019, 19, 274.
- Uzuner, Ö.; South, B.R.; Shen, S.; DuVall, S.L. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc. 2011, 18, 552–556.
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Li-Wei, H.L.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 1–9.
- Pradhan, S.; Elhadad, N.; South, B.R.; Martinez, D.; Christensen, L.; Vogel, A.; Suominen, H.; Chapman, W.W.; Savova, G. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J. Am. Med. Inform. Assoc. 2015, 22, 143–154.
- Vunikili, R.; SH, N.; Marica, G.; Farri, O. Clinical NER using Spanish BERT Embeddings. In Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), Malaga, Spain, 23 September 2020.
- Uzuner, O.; Bodnari, A.; Shen, S.; Forbush, T.; Pestian, J.; South, B.R. Evaluating the state of the art in coreference resolution for electronic medical records. J. Am. Med. Inform. Assoc. 2012, 19, 786–791.
- Sun, W.; Rumshisky, A.; Uzuner, O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J. Am. Med. Inform. Assoc. 2013, 20, 806–813.
- Henry, S.; Buchan, K.; Filannino, M.; Stubbs, A.; Uzuner, O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J. Am. Med. Inform. Assoc. 2019, 27, 3–12.
- Hand, D.J.; Christen, P.; Kirielle, N. F*: An interpretable transformation of the F-measure. Mach. Learn. 2021, 110, 451–456.
- Catelli, R.; Gargiulo, F.; Casola, V.; De Pietro, G.; Fujita, H.; Esposito, M. Crosslingual named entity recognition for clinical de-identification applied to a COVID-19 Italian data set. Appl. Soft Comput. 2020, 97, 106779.
- Nalluri, J.; Kapoor, R.; Sleeman, W.; Soni, P.; Ghosh, P.; Khajamoinuddin, S.; Hagan, M.; Palta, J. Health Information and Gateway Exchange (HINGE): Big Data Curation Tool for Radiation Oncology. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, E132.
- Aggarwal, A.; Mittal, M.; Battineni, G. Generative adversarial network: An overview of theory and applications. Int. J. Inf. Manag. Data Insights 2021, 1, 100004.
- Isaac, A.; Schlobach, S.; Matthezing, H.; Zinn, C. Integrated access to cultural heritage resources through representation and alignment of controlled vocabularies. Libr. Rev. 2008, 57, 187–199.
- Van Hooland, S.; Verborgh, R.; De Wilde, M.; Hercher, J.; Mannens, E.; Van de Walle, R. Evaluating the success of vocabulary reconciliation for cultural heritage collections. J. Am. Soc. Inf. Sci. Technol. 2013, 64, 464–479.
