Biofilm formation is an integral part of the microbial life cycle in nature. In food processing environments, bacterial transmissions occur primarily through raw or undercooked foods and by cross-contamination during unsanitary food preparation practices. Foodborne pathogens form biofilms as a survival strategy in various unfavorable environments, which also become a frequent source of recurrent contamination and outbreaks of foodborne illness.
- biofilm
- pathogenesis
- food safety
1. Introduction
Most microbes found in nature exist in biofilms, a well-structured, dynamic, diverse, synergistic and protective microbial community [1,2][1][2]. Biofilm formation (Figure 1) on a solid surface is a natural survival strategy of a microbial cell to compete efficiently with others for space and nutrients and to resist any unfavorable environmental conditions. The solid surface may be biotic (meat, produce, oral cavity, intestine, urogenital tract, skin, etc.) or abiotic (floors, walls, drains, equipment, or food-contacting surfaces). Microbes adhere to surfaces by producing an extracellular polymeric substance (EPS) forming a three-dimensional biofilm scaffold. Metaphorically, EPS is the “house” that covers and protects bacteria in biofilms [3]. Although biofilm architecture is solid, protecting bacteria from physical impact, most of the biofilm is still made up of water [4]. EPS makes up the majority of the total dry mass of biofilms. Approximately one-third of the biofilm’s dry weight is bacterial cells, and the remaining weight comes from bacteria-derived molecules, such as polysaccharides, proteins, and DNA, that make up the EPS [5,6][5][6]. Biofilms can be comprised of single-species or mixed-species cultures. The composition of bacteria in biofilms is also affected by surface materials, growth conditions, and biofilm maturity [7]. In the food processing environment, biofilm formation threatens food safety since pathogens can be directly transmitted through contact. After transmission, pathogens can also form biofilms on food surfaces. For instance, Listeria monocytogenes found on cantaloupe skin caused a multistate outbreak in 2011 [8,9][8][9].
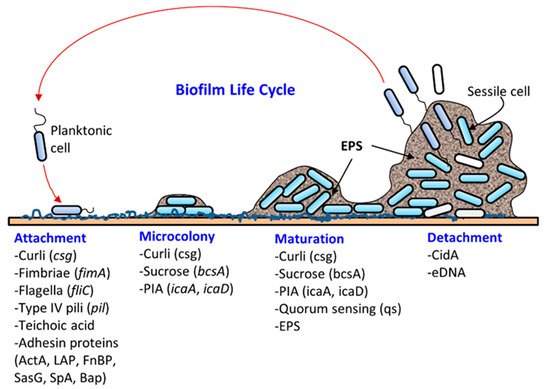
Microbial attachment and biofilm formation on solid surfaces provide the advantages of living in a protective scaffold against desiccation, antibiotics, or biocides (sanitizers), ultraviolet radiation, metallic cations, and physical impact from washing and cleaning. For instance, Martins et al. [10][11] recently showed that urinary tract infections caused by Staphylococcus saprophyticus were more resistant to several antibiotics in their biofilm status compared to their planktonic form. Likewise, biofilms of a commonly used model food spoilage bacterium Lactobacillus plantarum were more resistant to several biocides, including organic acids, ethanol, and sodium hypochlorite, than in its planktonic state [11][12]. Bacteria can acquire and/or exchange genetic materials in biofilms. DNA (plasmid) exchange can take place in biofilms through conjugation and transformation [12,13][13][14]. In addition, extracellular DNA can retain the electron shuttle molecule that is critical for redox cycling in biofilms [14][15].
Pathogen transmissions occur primarily through raw uncooked or undercooked foods and by cross-contamination during unsanitary food preparation practices. Pathogens find a harborage site or niche in food production facilities or product surfaces by forming biofilms [22][16]. These niches serve as a major source of foodborne outbreaks, especially in cafeterias, hospitals, cruise ships, and commercial food processing facilities. For example, the ubiquitous existence of L. monocytogenes in nature gives it numerous routes to be introduced in a food processing environment with various fresh produce or raw materials [23,24][17][18]. Once L. monocytogenes finds a niche in a food processing facility, it can attach to several abiotic surfaces, such as stainless steel, PVC, and polystyrene, and start to form biofilms, which can be resistant to sanitation and may lead to recurrent food contamination [25,26][19][20]. Repeated sampling of multiple food processing environments showed that similar L. monocytogenes strains can persist for a few months and up to 12 years [27][21]. The persistence of certain L. monocytogenes isolates in the food processing environment may also be due to the same strains that were consistently introduced by raw materials, or because of ineffective sanitation practices [28,29][22][23].
2. Bacterial Virulence Factors that Contribute to Biofilm Formation and Pathogenesis
Biofilm formation occurs in several stages: (i) attachment, (ii) microcolony formation, (iii) maturation with cellular differentiation, and (iv) detachment or dispersion ( Figure 1 ). In biofilms, microorganisms produce fimbriae, curli, flagella, adhesion proteins, and capsules to firmly attach to a surface [1,6][1][6]. Cells grow in close proximity and cell-to-cell communication (quorum sensing, QS) occurs through the production of autoinducers such as N-acyl homoserine lactone (AI-1) or other molecules, which also regulate gene expression for survival, growth, cell density, resistance to antimicrobials, tolerance to desiccation and pathogenesis [38,39][24][25]. Understanding the mechanism of quorum sensing in biofilm formation provides an opportunity for the application of appropriate QS inhibitors to control infection and pathogenesis [40,41,42,43,44][26][27][28][29][30]. As a microcolony continues to grow, cells accumulate forming a mature biofilm with three-dimensional scaffolding. Loose cells are then sloughed off from a mature biofilm and convert into planktonic cells, which start the life cycle of a biofilm again by attaching to new biotic and/or abiotic surfaces. The cells from biofilms could become a continuous source of food contamination [15][10]. Virulence factors that are involved in both biofilm formation and pathogenesis are discussed below for L. monocytogenes, Staphylococcus aureus, Escherichia coli, Salmonella enterica, and Pseudomonas aeruginosa.
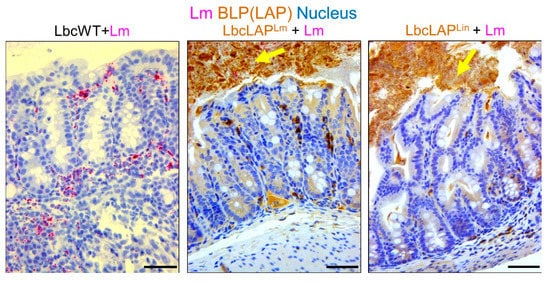
Listeria adhesion protein (LAP), an adhesion protein [49][31], has been indirectly attributed to the formation of biofilm ( Table 21 ). Our group has recently shown that recombinant Lactobacillus casei expressing LAP from L. monocytogenes or L. innocua on the bacterial surface showed aggregation and increased biofilm formation on a microtiter plate [51][32]. In a mouse model, these bioengineered strains also formed thicker biofilms on colonic villi than wild-type Lactobacillus casei ( Figure 32 ). Although the function of LAP in the pathogenesis of L. monocytogenes has been well documented [48[33][34],50], results from recombinant Lactobacillus casei highlights the role of LAP in biofilm formation as well.
| Bacteria | Factors | Function | Refs | |
|---|---|---|---|---|
| Biofilm Formation | Pathogenicity | |||
| Listeria monocytogenes | ActA (actin polymerization protein) | Bacterial sedimentation and aggregation | Rearrange host cytoskeletal structure and promote the cell-to-cell spread | [75][35] |
| LAP (Listeria adhesion protein) | Expression in recombinant Lactobacillus enhanced biofilm formation | Epithelial adhesion and translocation through the epithelial barrier | [50,51][34][32] | |
| PrfA (protein regulatory factor) | Regulate the expression of ActA that is necessary for biofilm formation | Regulatory protein that regulates the synthesis of multiple virulence factors | [71][36] | |
| WTA (wall teichoic acid) | Maintain cell wall (peptidoglycan) architecture and participate in biofilm formation | Induce inflammatory response | [77][37] | |
| Staphylococcus aureus | Bap (biofilm-associated protein) | Adhesion to inert surfaces and intercellular adhesion in the development of biofilm formation | Establish persistent infection on a mouse infection model | [101,103][38][39] |
| Protein A | Cell-to-cell adhesion in biofilm development; a major proteinaceous component in S. aureus biofilms | Help S. aureus to evade immune system in vivo | [99,100][40][41] | |
| PIA (polysaccharide intercellular adhesin) | Cell-to-cell binding in biofilm formation | Establish persistent in vivo infection | [91,93][42][43] | |
| Teichoic acid | Maintain cell wall (peptidoglycan) architecture and participate in biofilm formation | Induce inflammatory response | [76,77,111][44][37][45] | |
| FnBP (fibronectin-binding proteins) | Cell-to-cell adhesion through low-affinity homophilic interaction between neighboring cells | Promote bacterial attachment to host fibronectin for adhesion and colonization | [105,107][46][47] | |
| SasG (S. aureus surface protein G) | Zinc activated SasG-mediated biofilm formation | Adhesion to epithelial cells | [89,[48109]][49] | |
| Salmonella enterica | Fimbria (SEF17) | Cell-to-cell interaction in biofilm formation | Bind to human fibronectin and facilitate cell invasion | [144,145][50][51] |
| Bap (biofilm-associated protein) | Bap and curli can help form strong biofilms in both biotic and abiotic surface | Colonization, intestinal persistence, invasion to liver and spleen and lethality in mice | [126][52] | |
| CsgD, BcsA | Curli and cellulose synthesis | Colonization, biofilm formation and vertical transmission to egg | [146][53] | |
| Escherichia coli | Curli made with CsgA and CsgB | Adherence to abiotic surfaces | Adhere to epithelial cells when over expressed | [129,135][54][55] |
| Fim (fimbriae) | Biofilm formation on polystyrol | Adhesion to epithelial cell lines | [136][56] | |
| Enteroaggregative E. coli (EAEC) | Aggregative adherence fimbriae (AAF) | Mediate biofilm formation on abiotic surfaces | Bind to MUC1 on epithelial cells | [142,143][57][58] |
| Pseudomonas aeruginosa | PqsR | A key component of Pseudomonas quinolone signal system | Regulate the production of virulence factors, pyocyanin and hydrogen cyanide | [147][59] |
| Flagellum | Swimming motility and biofilm formation | Flagella is an important virulence factor. The flagellum-deficient strain showed less invasion in the mouse burn wound model and less colonization in the murine intestine | [148,149][60][61] | |
| Type IV pili | Twitching motility, and adhesion to abiotic surfaces | Adhesion to eukaryotic cells and pathogenesis | [150][62] | |
As one of the pathogens causing the most gastroenteritis cases around the world, E. coli is a model bacterium that forms biofilm after well-programmed production of various extracellular molecules [125][63]. Curli and cellulose are two major components making up the extracellular matrix [126][52].
In addition, in mixed-culture biofilms of P. aeruginosa and S. aureus, the presence of the latter organism can also increase exotoxin A expression [209,210][64][65], indicating that expression of virulence genes by one species in biofilms can be altered by the presence of another species.
3. Conclusions and Future Perspectives
In summary, multifunctional molecules involved in both bacterial pathogenesis and biofilm formation demonstrate a close connection between the two aspects. In L. monocytogenes, ActA rearranges actin in the host cell cytosol to propel cell-to-cell movement and also initiates biofilm formation by precipitating bacteria. Likewise, teichoic acids responsible for maintaining Gram-positive bacterial cell architecture also induce inflammatory response during infection and contribute to biofilm formation in both L. monocytogenes and S. aureus. Protein A of S. aureus not only helps the pathogen to evade the immune system but also facilitates cell-to-cell adhesion in biofilm development. Other proteins, including FnBP, SasG, and Bap, are also responsible for biofilm formation and pathogenesis in S. aureus. Curli is critical for biofilm formation and pathogenesis in E. coli. Similarly, curli and Bap are important in biofilm formation, intestinal colonization, and pathogenesis in gastroenteritis-causing non-typhoidal Salmonella. In Pseudomonas, PqsR plays a key role in the Pseudomonas quinolone signaling system and also regulates the production of virulence factors promoting bacterial biofilm formation and attachment to host epithelium. Other factors including flagella and type IV fimbriae are important in biofilm formation and colonization on epithelial cells. Many of the virulence factors that are involved in biofilm formation and host cell colonization have redundant functions, suggesting that even in the absence of one factor, bacteria can still form biofilms that are food safety and public health concerns.
Although the pathogenesis of multiple foodborne pathogens has been comprehensively studied, most of the results were generated using planktonic cultures under laboratory conditions. The actual risk of consuming pathogens from biofilms should be further characterized using animal models instead of only in vitro cultured mammalian cell models or virulence factor expression analyses. Recently, we used L. monocytogenes as a model foodborne pathogen to investigate the virulence of the bacteria in biofilms. Our data indicate that the virulence of biofilm-isolated L. monocytogenes was upregulated after 48 h bacterial adaption to the intestinal environment. These findings enhanced our understanding of bacterial pathogenesis of biofilm-isolated bacteria, and these data should be beneficial for the accurate evaluation of biofilm risks in food processing environments. Similarly, the assessment of the pathogenicity of other foodborne pathogens, such as E. coli and Salmonella, isolated from biofilms could also be further investigated using animal models. Using bacteria isolated from biofilms could also be a good model for studying bacteria switching from a saprophytic lifestyle to pathogenic status in animal hosts.
Although there are many studies of biofilm formation on plastic, stainless steel, or glass surfaces, more in-depth studies are needed of foodborne pathogen biofilms formed directly on food surfaces, for example, cantaloupe skin or eggshell. Bacteria isolated from these biofilms should represent a more realistic model to assess the risk of consuming foodborne pathogens found on food surfaces.
References
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633.
- Ren, D.; Madsen, J.S.; Sørensen, S.J.; Burmølle, M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015, 9, 81–89.
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947.
- Yaron, S.; Romling, U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 2014, 7, 496–516.
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322.
- Nadell, C.D.; Drescher, K.; Foster, K.R. Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 2016, 14, 589–600.
- Pinto, M.; Langer, T.M.; Hüffer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165.
- CDC, Multistate Outbreak of Listeriosis Linked to Whole Cantaloupes from Jensen Farms, Colorado (FINAL UPDATE). Available online: https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html (accessed on 27 August 2021).
- Fu, Y.; Deering, A.J.; Bhunia, A.K.; Yao, Y. Pathogen biofilm formation on cantaloupe surface and its impact on the antibacterial effect of lauroyl arginate ethyl. Food Microbiol. 2017, 64, 139–144.
- Ray, B.; Bhunia, A. Microbial attachements and biofilm formation. In Fundamental Food Microbiology, 5th ed.; Ray, B., Bhunia, A., Eds.; CRC Press: Boca Raton, FL, USA, 2014.
- Martins, K.B.; Ferreira, A.M.; Pereira, V.C.; Pinheiro, L.; Oliveira, A.D.; Cunha, M.D.L.R.D.S.D. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus saprophyticus isolated from patients with urinary tract infections. Front. Microbiol. 2019, 10, 40.
- Kubota, H.; Senda, S.; Tokuda, H.; Uchiyama, H.; Nomura, N. Stress resistance of biofilm and planktonic Lactobacillus plantarum subsp. plantarum JCM 1149. Food Microbiol. 2009, 26, 592–597.
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. Npj Biofilm. Microbiome 2016, 2, 1–5.
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195.
- Saunders, S.H.; Tse, E.C.M.; Yates, M.D.; Otero, F.J.; Trammell, S.A.; Stemp, E.D.A.; Barton, J.K.; Tender, L.M.; Newman, D.K. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 2020, 182, 919–932.e19.
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585.
- Ponniah, J.; Robin, T.; Paie, M.S.; Radu, S.; Ghazali, F.M.; Kqueen, C.Y.; Nishibuchi, M.; Nakaguchi, Y.; Malakar, P.K. Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Conrol 2010, 21, 774–778.
- Wu, S.; Wu, Q.; Zhang, J.; Chen, M.; Hu, H. Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS ONE 2015, 10, e0136682.
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561.
- Reis-Teixeira, F.B.D.; Alves, V.F.; Martinis, E.C.P.D. Growth, viability and architecture of biofilms of Listeria monocytogenes formed on abiotic surfaces. Brazilian J. Microbiol. 2017, 48, 587–591.
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8.
- Pan, Y.; Breidt, F.; Kathariou, S. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 2006, 72, 7711–7717.
- Langsrud, S.; Moen, B.; Møretrø, T.; Løype, M.; Heir, E. Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon-processing plants. J. Appl. Microbiol. 2016, 120, 366–378.
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823.
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.M.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189.
- Liu, Y.; Wu, L.; Han, J.; Dong, P.; Luo, X.; Zhang, Y.; Zhu, L. Inhibition of Biofilm Formation and Related Gene Expression of Listeria monocytogenes in Response to Four Natural Antimicrobial Compounds and Sodium Hypochlorite. Front. Microbiol. 2021, 11, 3523.
- Almeida, F.A.D.; Vargas, E.L.G.; Carneiro, D.G.; Pinto, U.M.; Vanetti, M.C.D. Virtual screening of plant compounds and nonsteroidal anti-inflammatory drugs for inhibition of quorum sensing and biofilm formation in Salmonella. Microb. Pathog. 2018, 121, 369–388.
- Aswathanarayan, J.B.; Vittal, R.R. Inhibition of biofilm formation and quorum sensing mediated phenotypes by berberine in Pseudomonas aeruginosa and Salmonella Typhimurium. RSC Adv. 2018, 8, 36133–36141.
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986.
- Kim, M.K.; Zhao, A.; Wang, A.; Brown, Z.Z.; Muir, T.W.; Stone, H.A.; Bassler, B.L. Surface-attached molecules control Staphylococcus aureus quorum sensing and biofilm development. Nat. Microbiol. 2017, 2, 17080.
- Jagadeesan, B.; Koo, O.K.; Kim, K.P.; Burkholder, K.M.; Mishra, K.K.; Aroonnual, A.; Bhunia, A.K. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 2010, 156, 2782–2795.
- Drolia, R.; Amalaradjou, M.A.R.; Ryan, V.; Tenguria, S.; Liu, D.; Bai, X.; Xu, L.; Singh, A.K.; Cox, A.D.; Bernal-Crespo, V.; et al. Receptor-targeted engineered probiotics mitigate lethal Listeria infection. Nat. Commun. 2020, 11, 6344.
- Burkholder, K.M.; Bhunia, A.K. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation, and induces expression of LAP receptor Hsp60. Infect. Immun. 2010, 78, 5062–5073.
- Drolia, R.; Tenguria, S.; Durkes, A.C.; Turner, J.R.; Bhunia, A.K. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe 2018, 23, 470–484.
- Travier, L.; Guadagnini, S.; Gouin, E.; Dufour, A.; Chenal-Francisque, V.; Cossart, P.; Olivo-Marin, J.-C.; Ghigo, J.-M.; Disson, O.; Lecuit, M. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013, 9, e1003131.
- Lemon, K.P.; Freitag, N.E.; Kolter, R. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J. Bacteriol. 2011, 192, 3969–3976.
- Zhu, X.; Liu, D.; Singh, A.K.; Drolia, R.; Bai, X.; Tenguria, S.; Bhunia, A.K. Tunicamycin mediated inhibition of wall teichoic acid affect Staphylococcus aureus and Listeria monocytogenes cell morphology, biofilm formation and virulence. Front. Microbiol. 2018, 9, 1352.
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, Í.; Penadés, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896.
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog. 2016, 12, e1005711.
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lopez, J.A.; Foster, T.J.; Penadés, J.R.; Lasa, I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843.
- Kobayashi, S.D.; DeLeo, F.R. Staphylococcus aureus protein A promotes immune suppression. mBio 2013, 4, e00764-13.
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1, 6-linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 1996, 178, 175–183.
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999, 67, 5427–5433.
- Gross, M.; Cramton, S.E.; Götz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426.
- Naclerio, G.A.; Onyedibe, K.I.; Sintim, H.O. Lipoteichoic Acid Biosynthesis Inhibitors as Potent Inhibitors of S. aureus and E. faecalis Growth and Biofilm Formation. Molecules 2020, 25, 2277.
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850.
- Gries, C.M.; Biddle, T.; Bose, J.L.; Kielian, T.; Lo, D.D. Staphylococcus aureus fibronectin binding protein A mediates biofilm development and infection. Infect. Immun. 2020, 88, e00859-19.
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446.
- Formosa-Dague, C.; Speziale, P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl. Acad. Sci. USA 2016, 113, 410–415.
- Austin, J.W.; Sanders, G.; Kay, W.W.; Collinson, S.K. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 1998, 162, 295–301.
- Collinson, S.; Doig, P.; Doran, J.; Clouthier, S.; Kay, W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 1993, 175, 12–18.
- Latasa, C.; Roux, A.; Toledo-Arana, A.; Ghigo, J.M.; Gamazo, C.; Penadés, J.R.; Lasa, I. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 2005, 58, 1322–1339.
- Chen, S.; Feng, Z.; Sun, H.; Zhang, R.; Qin, T.; Peng, D. Biofilm-Formation-Related Genes csgD and bcsA Promote the Vertical Transmission of Salmonella Enteritidis in Chicken. Front. Vet. Sci. 2020, 7, 625049.
- Nguyen, P.Q.; Botyanszki, Z.; Tay, P.K.R.; Joshi, N.S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 2014, 5, 1–10.
- Saldaña, Z.; Xicohtencatl-Cortes, J.; Avelino, F.; Phillips, A.D.; Kaper, J.B.; Puente, J.L.; Girón, J.A. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 2009, 11, 992–1006.
- Elpers, L.; Hensel, M. Expression and functional characterization of various chaperon-usher fimbriae, curli fimbriae, and type 4 pili of enterohemorrhagic Escherichia coli O157:H7 Sakai. Front. Microbiol. 2020, 11, 378.
- Sheikh, J.; Hicks, S.; Dall’Agnol, M.; Phillips, A.D.; Nataro, J.P. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 2001, 41, 983–997.
- Boll, E.J.; Ayala-Lujan, J.; Szabady, R.L.; Louissaint, C.; Smith, R.Z.; Krogfelt, K.A.; Nataro, J.P.; Ruiz-Perez, F.; McCormick, B.A. Enteroaggregative Escherichia coli adherence fimbriae drive inflammatory cell recruitment via interactions with epithelial MUC1. mBio 2017, 8, e00717-17.
- Farrow, J.M.; Sund, Z.M.; Ellison, M.L.; Wade, D.S.; Coleman, J.P.; Pesci, E.C. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J. Bacteriol. 2008, 190, 7043–7051.
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524.
- Pier, G.B.; Meluleni, G.; Goldberg, J.B. Clearance of Pseudomonas aeruginosa from the murine gastrointestinal tract is effectively mediated by O-antigen-specific circulating antibodies. Infect. Immun. 1995, 63, 2818–2825.
- Déziel, E.; Comeau, Y.; Villemur, R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 2001, 183, 1195–1204.
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 249–289.
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004.
- Goldsworthy, M.J.H. Gene expression of Pseudomonas aeruginosa and MRSA within a catheter-associated urinary tract infection biofilm model. BioSci. Horizon 2008, 1, 28–37.
