The burden of fungal infections is not widely appreciated. Although these infections are responsible for over one million deaths annually, it is estimated that one billion people are affected by severe fungal diseases. Mycoses of nails and skin, primarily caused by fungi known as dermatophytes, are the most common fungal infections. Trichophyton rubrum appears to be the most common causative agent of dermatophytosis, followed by Trichophyton interdigitale. An estimated 25% of the world’s population suffers from dermatomycosis. Although these infections are not lethal, they compromise the quality of life of infected patients. The outcome of antidermatophytic treatments is impaired by various conditions, such as resistance and tolerance of certain dermatophyte strains.
- Trichophyton
- dermatophytosis
- cutaneous infection
- fungal-host interaction
- virulence traits
- transcription factor
- resistance mechanisms
- keratin degradation
1. Introduction
2. Virulence Attributes of Dermatophytes
2.1. Infection Models and Fungal-Host Interaction
2.2. Transcription Factors and Fungal Signaling Pathways as Virulence Traits
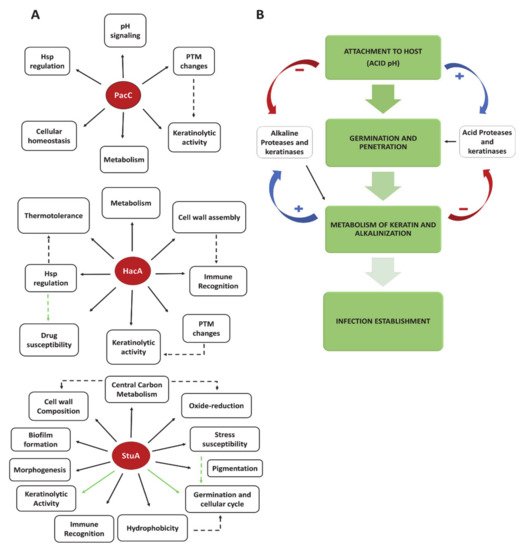
3. Treatment, Clinical Implications, and Perspectives
3.1. Therapeutic Options and Resistance Mechanisms
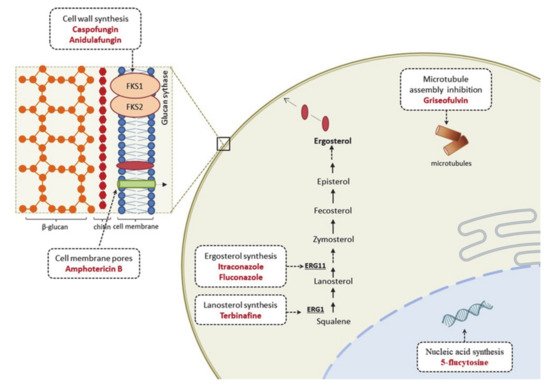
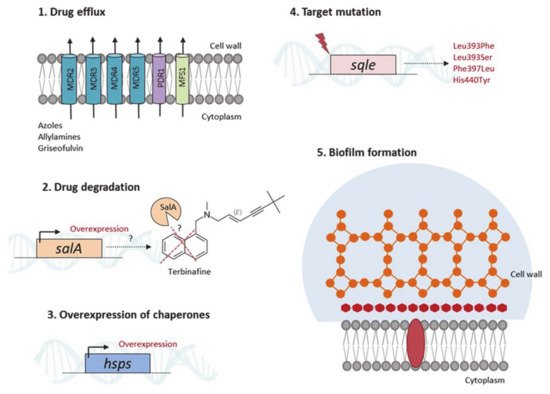
3.2. From Knowing the Enemy to Identifying Novel Antifungal Targets
References
- De Hoog, S.; Monod, M.; Dawson, T.; Boekhout, T.; Mayser, P.; Gräser, Y. Skin Fungi from Colonization to Infection. Microbiol. Spectr. 2017, 5.
- Mushtaq, S.; Faizi, N.; Amin, S.S.; Adil, M.; Mohtashim, M. Impact on quality of life in patients with dermatophytosis. Australas. J. Dermatol. 2020, 61, e184–e188.
- Narang, T.; Bhattacharjee, R.; Singh, S.; Jha, K.; Kavita, K.; Mahajan, R.; Dogra, S. Quality of life and psychological morbidity in patients with superficial cutaneous dermatophytosis. Mycoses 2019, 62, 680–685.
- Kovitwanichkanont, T.; Chong, A.H. Superficial fungal infections. Aust. J. Gen. Pract. 2019, 48, 706–711.
- Dogra, S.; Narang, T. Emerging atypical and unusual presentations of dermatophytosis in India. Clin. Dermatol. Rev. 2017, 1, 12.
- Atzori, L.; Pau, M.; Aste, N.; Aste, N. Dermatophyte infections mimicking other skin diseases: A 154-person case survey of tinea atypica in the district of Cagliari (Italy). Int. J. Dermatol. 2012, 51, 410–415.
- Boral, H.; Durdu, M.; Ilkit, M. Infection and Drug Resistance Majocchi’s granuloma: Current perspectives. IDR 2018.
- Rouzaud, C.; Hay, R.; Chosidow, O.; Dupin, N.; Puel, A.; Lortholary, O.; Lanternier, F. Severe Dermatophytosis and Acquired or Innate Immunodeficiency: A Review. J. Fungi 2015, 2, 4.
- Agarwal, A.; Hassanandani, T.; Das, A.; Panda, M.; Chakravorty, S. “Mask tinea”: Tinea faciei possibly potentiated by prolonged mask usage during the COVID-19 pandemic. Clin. Exp. Dermatol. 2021, 46, 190–193.
- Gnat, S.; Nowakiewicz, A.; Łagowski, D.; Zięba, P. Host- and pathogen-dependent susceptibility and predisposition to dermatophytosis. J. Med. Microbiol. 2019, 68, 823–836.
- Gnat, S.; Łagowski, D.; Nowakiewicz, A. Genetic Predisposition and Its Heredity in the Context of Increased Prevalence of Dermatophytoses. Mycopathologia 2021, 186, 163–176.
- Lanternier, F.; Pathan, S.; Vincent, Q.B.; Liu, L.; Cypowyj, S.; Prando, C.; Migaud, M.; Taibi, L.; Ammar-Khodja, A.; Stambouli, O.B.; et al. Deep dermatophytosis and inherited CARD9 deficiency. N. Engl. J. Med. 2013, 369, 1704–1714.
- Begum, J.; Mir, N.A.; Lingaraju, M.C.; Buyamayum, B.; Dev, K. Recent advances in the diagnosis of dermatophytosis. J. Basic Microbiol. 2020, 60, 293–303.
- De Hoog, G.S.; Dukik, K.; Monod, M.; Packeu, A.; Stubbe, D.; Hendrickx, M.; Kupsch, C.; Stielow, J.B.; Freeke, J.; Göker, M.; et al. Toward a Novel Multilocus Phylogenetic Taxonomy for the Dermatophytes. Mycopathologia 2017, 182, 5–31.
- Verma, S.B.; Panda, S.; Nenoff, P.; Singal, A.; Rudramurthy, S.M.; Uhrlass, S.; Das, A.; Bisherwal, K.; Shaw, D.; Vasani, R. The unprecedented epidemic-like scenario of dermatophytosis in India: I. Epidemiology, risk factors and clinical features. Indian J. Dermatol. Venereol. Leprol. 2021, 87, 154–175.
- Gupta, A.K.; Stec, N.; Summerbell, R.C.; Shear, N.H.; Piguet, V.; Tosti, A.; Piraccini, B.M. Onychomycosis: A review. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1972–1990.
- Rodríguez-Cerdeira, C.; Martínez-Herrera, E.; Szepietowski, J.C.; Pinto-Almazán, R.; Frías-De-León, M.G.; Espinosa-Hernández, V.M.; Chávez-Gutiérrez, E.; García-Salazar, E.; Vega-Sánchez, D.C.; Arenas, R.; et al. A systematic review of worldwide data on tinea capitis: Analysis of the last 20 years. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 844–883.
- Sacheli, R.; Harag, S.; Dehavay, F.; Evrard, S.; Rousseaux, D.; Adjetey, A.; Seidel, L.; Laffineur, K.; Lagrou, K.; Hayette, M.-P. Belgian National Survey on Tinea Capitis: Epidemiological Considerations and Highlight of Terbinafine-Resistant T. mentagrophytes with a Mutation on SQLE Gene. J. Fungi 2020, 6, 195.
- Bontems, O.; Fratti, M.; Salamin, K.; Guenova, E.; Monod, M. Epidemiology of Dermatophytoses in Switzerland According to a Survey of Dermatophytes Isolated in Lausanne between 2001 and 2018. J. Fungi 2020, 6, 95.
- Wang, R.; Huang, C.; Zhang, Y.; Li, R. Invasive dermatophyte infection: A systematic review. Mycoses 2021, 64, 340–348.
- Maranhão, F.C.A.; Paião, F.G.; Martinez-Rossi, N.M. Isolation of transcripts over-expressed in human pathogen Trichophyton rubrum during growth in keratin. Microb. Pathog. 2007, 43, 166–172.
- Maranhão, F.C.A.; Silveira, H.C.S.; Rossi, A.; Martinez-Rossi, N.M. Isolation of transcripts overexpressed in the human pathogen Trichophyton rubrum grown in lipid as carbon source. Can. J. Microbiol. 2011, 57, 333–338.
- Staib, P.; Zaugg, C.; Mignon, B.; Weber, J.; Grumbt, M.; Pradervand, S.; Harshman, K.; Monod, M. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology 2010, 156, 884–895.
- De Faria, L.V.; do Carmo, P.H.F.; da Costa, M.C.; Peres, N.T.A.; Rodrigues Chagas, I.A.; Furst, C.; Ferreira, G.F.; Costa, A.O.; Santos, D.A. Acanthamoeba castellanii as an alternative interaction model for the dermatophyte Trichophyton rubrum. Mycoses 2020, 63, 1331–1340.
- Campos, M.R.M.; Russo, M.; Gomes, E.; Almeida, S.R. Stimulation, inhibition and death of macrophages infected with Trichophyton rubrum. Microbes Infect. 2006, 8, 372–379.
- Heddergott, C.; Bruns, S.; Nietzsche, S.; Leonhardt, I.; Kurzai, O.; Kniemeyer, O.; Brakhage, A.A. The Arthroderma benhamiae hydrophobin HypA mediates hydrophobicity and influences recognition by human immune effector cells. Eukaryot. Cell 2012, 11, 673–682.
- Faway, É.; Lambert de Rouvroit, C.; Poumay, Y. In vitro models of dermatophyte infection to investigate epidermal barrier alterations. Exp. Dermatol. 2018, 27, 915–922.
- Ishii, M.; Matsumoto, Y.; Yamada, T.; Abe, S.; Sekimizu, K. An invertebrate infection model for evaluating anti-fungal agents against dermatophytosis. Sci. Rep. 2017, 7, 12289.
- Baltazar, L.; de, M.; Santos, P.C.; de Paula, T.P.; Rachid, M.A.; Cisalpino, P.S.; Souza, D.G.; Santos, D.A. IFN-γ impairs Trichophyton rubrum proliferation in a murine model of dermatophytosis through the production of IL-1β and reactive oxygen species. Med. Mycol. 2014, 52, 293–302.
- Peres, N.T.A.; Silva, L.G.; Santos, R.S.; Jacob, T.R.; Persinoti, G.F.; Rocha, L.B.; Falcão, J.P.; Rossi, A.; Martinez-Rossi, N.M. In vitro and ex vivo infection models help assess the molecular aspects of the interaction of Trichophyton rubrum with the host milieu. Med. Mycol. 2016, 54, 420–427.
- Johns, L.E.; Goldman, G.H.; Ries, L.N.A.; Brown, N.A. Nutrient sensing and acquisition in fungi: Mechanisms promoting pathogenesis in plant and human hosts. Fungal Biol. Rev. 2021, 36, 1–14.
- Kaufman, G.; Horwitz, B.A.; Duek, L.; Ullman, Y.; Berdicevsky, I. Infection stages of the dermatophyte pathogen Trichophyton: Microscopic characterization and proteolytic enzymes. Med. Mycol. 2007, 45, 149–155.
- Tainwala, R.; Sharma, Y. Pathogenesis of dermatophytoses. Indian J. Dermatol. 2011, 56, 259–261.
- Baldo, A.; Mathy, A.; Tabart, J.; Camponova, P.; Vermout, S.; Massart, L.; Maréchal, F.; Galleni, M.; Mignon, B. Secreted subtilisin Sub3 from Microsporum canis is required for adherence to but not for invasion of the epidermis. Br. J. Dermatol. 2010, 162, 990–997.
- Verstrepen, K.J.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990.
- De Abreu, M.H.; Bitencourt, T.A.; Franco, M.E.; Moreli, I.S.; Cantelli, B.A.M.; Komoto, T.T.; Marins, M.; Fachin, A.L. Expression of genes containing tandem repeat patterns involved in the fungal-host interaction and in the response to antifungals in Trichophyton rubrum. Mycoses 2020, 63, 610–616.
- Bitencourt, T.A.; Macedo, C.; Franco, M.E.; Assis, A.F.; Komoto, T.T.; Stehling, E.G.; Beleboni, R.O.; Malavazi, I.; Marins, M.; Fachin, A.L. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genom. 2016, 17, 249.
- Lopes, L.; Bitencourt, T.A.; Lang, E.A.S.; Sanches, P.R.; Peres, N.T.A.; Rossi, A.; Martinez-Rossi, N.M. Genes coding for LysM domains in the dermatophyte Trichophyton rubrum: A transcription analysis. Med. Mycol. 2020, 58, 372–379.
- Kar, B.; Patel, P.; Free, S.J. Trichophyton rubrum LysM proteins bind to fungal cell wall chitin and to the N-linked oligosaccharides present on human skin glycoproteins. PLoS ONE 2019, 14, e0215034.
- Lang, E.A.S.; Bitencourt, T.A.; Peres, N.T.A.; Lopes, L.; Silva, L.G.; Cazzaniga, R.A.; Rossi, A.; Martinez-Rossi, N.M. The stuA gene controls development, adaptation, stress tolerance, and virulence of the dermatophyte Trichophyton rubrum. Microbiol. Res. 2020, 241, 126592.
- Martins, M.P.; Silva, L.G.; Rossi, A.; Sanches, P.R.; Souza, L.D.R.; Martinez-Rossi, N.M. Global Analysis of Cell Wall Genes Revealed Putative Virulence Factors in the Dermatophyte Trichophyton rubrum. Front. Microbiol. 2019, 10, 2168.
- Bitencourt, T.A.; Rezende, C.P.; Quaresemin, N.R.; Moreno, P.; Hatanaka, O.; Rossi, A.; Martinez-Rossi, N.M.; Almeida, F. Extracellular Vesicles from the Dermatophyte Trichophyton interdigitale Modulate Macrophage and Keratinocyte Functions. Front. Immunol. 2018, 9, 2343.
- Leng, W.; Liu, T.; Li, R.; Yang, J.; Wei, C.; Zhang, W.; Jin, Q. Proteomic profile of dormant Trichophyton rubrum conidia. BMC Genom. 2008, 9, 303.
- De Peres, N.T.A.; Maranhão, F.C.A.; Rossi, A.; Martinez-Rossi, N.M. Dermatophytes: Host-pathogen interaction and antifungal resistance. An. Bras. Dermatol. 2010, 85, 657–667.
- Grumbt, M.; Monod, M.; Yamada, T.; Hertweck, C.; Kunert, J.; Staib, P. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J. Invest. Dermatol. 2013, 133, 1550–1555.
- Kasperova, A.; Kunert, J.; Raska, M. The possible role of dermatophyte cysteine dioxygenase in keratin degradation. Med. Mycol. 2013, 51, 449–454.
- Ciesielska, A.; Kawa, A.; Kanarek, K.; Soboń, A.; Szewczyk, R. Metabolomic analysis of Trichophyton rubrum and Microsporum canis during keratin degradation. Sci. Rep. 2021, 11, 3959.
- Martins, M.P.; Rossi, A.; Sanches, P.R.; Bortolossi, J.C.; Martinez-Rossi, N.M. Comprehensive analysis of the dermatophyte Trichophyton rubrum transcriptional profile reveals dynamic metabolic modulation. Biochem. J. 2020, 477, 873–885.
- Grumbt, M.; Defaweux, V.; Mignon, B.; Monod, M.; Burmester, A.; Wöstemeyer, J.; Staib, P. Targeted gene deletion and in vivo analysis of putative virulence gene function in the pathogenic dermatophyte Arthroderma benhamiae. Eukaryot. Cell 2011, 10, 842–853.
- Martinez-Rossi, N.M.; Peres, N.T.A.; Rossi, A. Pathogenesis of Dermatophytosis: Sensing the Host Tissue. Mycopathologia 2017, 182, 215–227.
- Lambers, H.; Piessens, S.; Bloem, A.; Pronk, H.; Finkel, P. Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int. J. Cosmet. Sci. 2006, 28, 359–370.
- Wertz, P.W.; Szalay, S. Innate Antimicrobial Defense of Skin and Oral Mucosa. Antibiotics 2020, 9, 159.
- Martinez-Rossi, N.M.; Persinoti, G.F.; Peres, N.T.A.; Rossi, A. Role of pH in the pathogenesis of dermatophytoses. Mycoses 2012, 55, 381–387.
- Peres, N.T.A.; Sanches, P.R.; Falco, J.P.; Silveira, H.C.S.; Paião, F.G.; Maranhão, F.C.A.; Gras, D.E.; Segato, F.; Cazzaniga, R.A.; Mazucato, M.; et al. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 2010, 10, 39.
- Cornet, M.; Gaillardin, C. pH signaling in human fungal pathogens: A new target for antifungal strategies. Eukaryot. Cell 2014, 13, 342–352.
- Ferreira-Nozawa, M.S.; Silveira, H.C.S.; Ono, C.J.; Fachin, A.L.; Rossi, A.; Martinez-Rossi, N.M. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med. Mycol. 2006, 44, 641–645.
- Persinoti, G.F.; Martinez, D.A.; Li, W.; Döğen, A.; Billmyre, R.B.; Averette, A.; Goldberg, J.M.; Shea, T.; Young, S.; Zeng, Q.; et al. Whole-Genome Analysis Illustrates Global Clonal Population Structure of the Ubiquitous Dermatophyte Pathogen Trichophyton rubrum. Genetics 2018, 208, 1657–1669.
- Silveira, H.C.S.; Gras, D.E.; Cazzaniga, R.A.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. Transcriptional profiling reveals genes in the human pathogen Trichophyton rubrum that are expressed in response to pH signaling. Microb. Pathog. 2010, 48, 91–96.
- Díez, E.; Alvaro, J.; Espeso, E.A.; Rainbow, L.; Suárez, T.; Tilburn, J.; Arst, H.N.; Peñalva, M.A. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002, 21, 1350–1359.
- Peñas, M.M.; Hervás-Aguilar, A.; Múnera-Huertas, T.; Reoyo, E.; Peñalva, M.A.; Arst, H.N.; Tilburn, J. Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot. Cell 2007, 6, 960–970.
- Rossi, A.; Cruz, A.H.S.; Santos, R.S.; Silva, P.M.; Silva, E.M.; Mendes, N.S.; Martinez-Rossi, N.M. Ambient pH sensing in filamentous fungi: Pitfalls in elucidating regulatory hierarchical signaling networks. IUBMB Life 2013, 65, 930–935.
- Martins, M.P.; Martinez-Rossi, N.M.; Sanches, P.R.; Rossi, A. The PAC-3 transcription factor critically regulates phenotype-associated genes in Neurospora crassa. Genet. Mol. Biol. 2020, 43, e20190374.
- Mendes, N.S.; Trevisan, G.L.; Silva Cruz, A.H.; Santos, R.S.; Peres, N.T.A.; Martinez-Rossi, N.M.; Rossi, A. Transcription of N- and O-linked mannosyltransferase genes is modulated by the pacC gene in the human dermatophyte Trichophyton rubrum. FEBS Open Bio 2012, 2, 294–297.
- Nozawa, S.R.; Ferreira-Nozawa, M.S.; Martinez-Rossi, N.M.; Rossi, A. The pH-induced glycosylation of secreted phosphatases is mediated in Aspergillus nidulans by the regulatory gene pacC-dependent pathway. Fungal Genet. Biol. 2003, 39, 286–295.
- Nozawa, S.R.; Thede, G., Jr.; Crott, L.S.P.; Barbosa, J.E.; Rossi, A. The synthesis of Phosphate-repressible alkaline phosphatase do not appear to be regulated by ambient pH in the filamentous mould Neurospora crassa. Braz. J. Microbiol. 2002, 33, 92–95.
- Da Silva, L.G.; Martins, M.P.; Sanches, P.R.; de Peres, N.T.A.; Martinez-Rossi, N.M.; Rossi, A. Saline stress affects the pH-dependent regulation of the transcription factor PacC in the dermatophyte Trichophyton interdigitale. Braz. J. Microbiol. 2020, 51, 1585–1591.
- Römisch, K. A cure for traffic jams: Small molecule chaperones in the endoplasmic reticulum. Traffic 2004, 5, 815–820.
- Moore, K.A.; Hollien, J. The unfolded protein response in secretory cell function. Annu. Rev. Genet. 2012, 46, 165–183.
- Saloheimo, M.; Valkonen, M.; Penttilä, M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 2003, 47, 1149–1161.
- Bitencourt, T.A.; Lang, E.A.S.; Sanches, P.R.; Peres, N.T.A.; Oliveira, V.M.; Fachin, A.L.; Rossi, A.; Martinez-Rossi, N.M. HacA Governs Virulence Traits and Adaptive Stress Responses in Trichophyton rubrum. Front. Microbiol. 2020, 11, 193.
- Wiederrecht, G.; Seto, D.; Parker, C.S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 1988, 54, 841–853.
- Martínez-Pastor, M.T.; Marchler, G.; Schüller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235.
- Boy-Marcotte, E.; Lagniel, G.; Perrot, M.; Bussereau, F.; Boudsocq, A.; Jacquet, M.; Labarre, J. The heat shock response in yeast: Differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 1999, 33, 274–283.
- Neves-da-Rocha, J.; Bitencourt, T.A.; de Oliveira, V.M.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. Alternative Splicing in Heat Shock Protein Transcripts as a Mechanism of Cell Adaptation in Trichophyton rubrum. Cells 2019, 8, 1206.
- Jacob, T.R.; Peres, N.T.A.; Martins, M.P.; Lang, E.A.S.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. Heat Shock Protein 90 (Hsp90) as a Molecular Target for the Development of Novel Drugs Against the Dermatophyte Trichophyton rubrum. Front. Microbiol. 2015, 6, 1241.
- Martinez-Rossi, N.M.; Jacob, T.R.; Sanches, P.R.; Peres, N.T.A.; Lang, E.A.S.; Martins, M.P.; Rossi, A. Heat Shock Proteins in Dermatophytes: Current Advances and Perspectives. Curr. Genom. 2016, 17, 99–111.
- Bitencourt, T.A.; Neves-da-Rocha, J.; Martins, M.P.; Sanches, P.R.; Lang, E.A.S.; Bortolossi, J.C.; Rossi, A.; Martinez-Rossi, N.M. StuA-regulated processes in the dermatophyte Trichophyton rubrum: Transcription profile, cell-cell adhesion, and immunomodulation. Front. Cell. Infect. Microbiol. 2021, 11, 447.
- Kröber, A.; Etzrodt, S.; Bach, M.; Monod, M.; Kniemeyer, O.; Staib, P.; Brakhage, A.A. The transcriptional regulators SteA and StuA contribute to keratin degradation and sexual reproduction of the dermatophyte Arthroderma benhamiae. Curr. Genet. 2017, 63, 103–116.
- Liu, H.; Xu, W.; Bruno, V.M.; Phan, Q.T.; Solis, N.V.; Woolford, C.A.; Ehrlich, R.L.; Shetty, A.C.; McCraken, C.; Lin, J.; et al. Determining Aspergillus fumigatus transcription factor expression and function during invasion of the mammalian lung. PLoS Pathog. 2021, 17, e1009235.
- Badali, H.; Mohammadi, R.; Mashedi, O.; de Hoog, G.S.; Meis, J.F. In vitro susceptibility patterns of clinically important Trichophyton and Epidermophyton species against nine antifungal drugs. Mycoses 2015, 58, 303–307.
- Salehi, Z.; Fatahi, N.; Taran, M.; Izadi, A.; Badali, H.; Hashemi, S.J.; Rezaie, S.; Daie Ghazvini, R.; Ghaffari, M.; Aala, F.; et al. Comparison of in vitro antifungal activity of novel triazoles with available antifungal agents against dermatophyte species caused tinea pedis. J. Mycol. Med. 2020, 30, 100935.
- Chen, X.; Jiang, X.; Yang, M.; Bennett, C.; González, U.; Lin, X.; Hua, X.; Xue, S.; Zhang, M. Systemic antifungal therapy for tinea capitis in children: An abridged Cochrane Review. J. Am. Acad. Dermatol. 2017, 76, 368–374.
- Tey, H.L.; Tan, A.S.L.; Chan, Y.C. Meta-analysis of randomized, controlled trials comparing griseofulvin and terbinafine in the treatment of tinea capitis. J. Am. Acad. Dermatol. 2011, 64, 663–670.
- Khurana, A.; Sardana, K.; Chowdhary, A. Antifungal resistance in dermatophytes: Recent trends and therapeutic implications. Fungal Genet. Biol. 2019, 132, 103255.
- Ebert, A.; Monod, M.; Salamin, K.; Burmester, A.; Uhrlaß, S.; Wiegand, C.; Hipler, U.; Krüger, C.; Koch, D.; Wittig, F.; et al. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: A multicentre study. Mycoses 2020, 63, 717–728.
- Martinez-Rossi, N.M.; Peres, N.T.A.; Rossi, A. Antifungal Resistance Mechanisms in Dermatophytes. Mycopathologia 2008, 166, 369–383.
- Martins, M.P.; Rossi, A.; Sanches, P.R.; Martinez-Rossi, N.M. Differential expression of multidrug-resistance genes in Trichophyton rubrum. J. Integr. OMICS 2019, 9, 65–69.
- Yamada, T.; Maeda, M.; Alshahni, M.M.; Tanaka, R.; Yaguchi, T.; Bontems, O.; Salamin, K.; Fratti, M.; Monod, M. Terbinafine Resistance of Trichophyton Clinical Isolates Caused by Specific Point Mutations in the Squalene Epoxidase Gene. Antimicrob. Agents Chemother. 2017, 61, e00115-17.
- Saunte, D.M.L.; Hare, R.K.; Jørgensen, K.M.; Jørgensen, R.; Deleuran, M.; Zachariae, C.O.; Thomsen, S.F.; Bjørnskov-Halkier, L.; Kofoed, K.; Arendrup, M.C. Emerging Terbinafine Resistance in Trichophyton: Clinical Characteristics, Squalene Epoxidase Gene Mutations, and a Reliable EUCAST Method for Detection. Antimicrob. Agents Chemother. 2019, 63, e01126-19.
- Singh, A.; Masih, A.; Khurana, A.; Singh, P.K.; Gupta, M.; Hagen, F.; Meis, J.F.; Chowdhary, A. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses 2018, 61, 477–484.
- Kano, R.; Kimura, U.; Kakurai, M.; Hiruma, J.; Kamata, H.; Suga, Y.; Harada, K. Trichophyton indotineae sp. nov.: A New Highly Terbinafine-Resistant Anthropophilic Dermatophyte Species. Mycopathologia 2020, 185, 947–958.
- Tang, C.; Kong, X.; Ahmed, S.; Thakur, R.; Chowdhary, A.; Nenoff, P.; Uhrlass, S.; Verma, S.; Meis, J.; Kandemir, H.; et al. Taxonomy of the Trichophyton mentagrophytes/T. interdigitale Species Complex Harboring the Highly Virulent, Multiresistant Genotype T. indotineae. Mycopathologia 2021, 186, 315–326.
- Santos, H.L.; Lang, E.A.S.; Segato, F.; Rossi, A.; Martinez-Rossi, N.M. Terbinafine resistance conferred by multiple copies of the salicylate 1-monooxygenase gene in Trichophyton rubrum. Med. Mycol. 2018, 56, 378–381.
- Monod, M.; Feuermann, M.; Salamin, K.; Fratti, M.; Makino, M.; Alshahni, M.M.; Makimura, K.; Yamada, T. Trichophyton rubrum Azole Resistance Mediated by a New ABC Transporter, TruMDR3. Antimicrob. Agents Chemother. 2019, 63, e00863-19.
- Cervelatti, E.P.; Fachin, A.L.; Ferreira-Nozawa, M.S.; Martinez-Rossi, N.M. Molecular cloning and characterization of a novel ABC transporter gene in the human pathogen Trichophyton rubrum. Med. Mycol. 2006, 44, 141–147.
- Martins, M.P.; Franceschini, A.C.C.; Jacob, T.R.; Rossi, A.; Martinez-Rossi, N.M. Compensatory expression of multidrug-resistance genes encoding ABC transporters in dermatophytes. J. Med. Microbiol. 2016, 65, 605–610.
- Fachin, A.L.; Ferreira-Nozawa, M.S.; Maccheroni, W.; Martinez-Rossi, N.M. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J. Med. Microbiol. 2006, 55, 1093–1099.
- De Castelo-Branco, D.S.C.M.; de Aguiar, L.; Araújo, G.D.S.; Lopes, R.G.P.; de Sales, J.A.; Pereira-Neto, W.A.; de Pinheiro, A.Q.; Paixão, G.C.; de Cordeiro, R.A.; Sidrim, J.J.C.; et al. In vitro and ex vivo biofilms of dermatophytes: A new panorama for the study of antifungal drugs. Biofouling 2020, 36, 783–791.
- Peres, N.T.A.; Cursino-Santos, J.R.; Rossi, A.; Martinez-Rossi, N.M. In vitro susceptibility to antimycotic drug undecanoic acid, a medium-chain fatty acid, is nutrient-dependent in the dermatophyte Trichophyton rubrum. World J. Microbiol. Biotechnol. 2011, 27, 1719–1723.
- Lin, H.; Liu, X.; Shen, Z.; Cheng, W.; Zeng, Z.; Chen, Y.; Tang, C.; Jiang, T. The effect of isoflavaspidic acid PB extracted from Dryopteris fragrans (L.) Schott on planktonic and biofilm growth of dermatophytes and the possible mechanism of antibiofilm. J. Ethnopharmacol. 2019, 241, 111956.
- Deng, S.; Zhang, C.; Seyedmousavi, S.; Zhu, S.; Tan, X.; Wen, Y.; Huang, X.; Lei, W.; Zhou, Z.; Fang, W.; et al. Comparison of the in vitro activities of newer triazoles and established antifungal agents against Trichophyton rubrum. Antimicrob. Agents Chemother. 2015, 59, 4312–4314.
- Hur, M.S.; Park, M.; Jung, W.H.; Lee, Y.W. Evaluation of drug susceptibility test for Efinaconazole compared with conventional antifungal agents. Mycoses 2019, 62, 291–297.
- Roemer, T.; Krysan, D.J. Antifungal drug development: Challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 2014, 4, a019703.
- Yamada, T.; Makimura, K.; Abe, S. Isolation, characterization, and disruption of dnr1, the areA/nit-2-like nitrogen regulatory gene of the zoophilic dermatophyte, Microsporum canis. Med. Mycol. 2006, 44, 243–252.
- Yamada, T.; Makimura, K.; Satoh, K.; Umeda, Y.; Ishihara, Y.; Abe, S. Agrobacterium tumefaciens-mediated transformation of the dermatophyte, Trichophyton mentagrophytes: An efficient tool for gene transfer. Med. Mycol. 2009, 47, 485–494.
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656.
- Mendes, N.S.; Bitencourt, T.A.; Sanches, P.R.; Silva-Rocha, R.; Martinez-Rossi, N.M.; Rossi, A. Transcriptome-wide survey of gene expression changes and alternative splicing in Trichophyton rubrum in response to undecanoic acid. Sci. Rep. 2018, 8, 2520.
- Gomes, E.; Bortolossi, J.; Sanches, P.; Mendes, N.; Martinez-Rossi, N.; Rossi, A. STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum. Int. J. Mol. Sci. 2018, 19, 3654.
