Salt-tolerant Plant Growth-Promoting Rhizobacteria (PGPR) has been reported to possess various salt-tolerant mechanisms, namely through the production of exopolysaccharide (EPS) and the formation of a biofilm, which have been proven to restrict Na+ uptake under saline soil conditions.
Salt-tolerant plant growth-promoting rhizobacteria (PGPR) could be an alternative to
alleviate salinity problems in rice plants grown in the coastal areas. This study was conducted
to isolate and characterize salt-tolerant PGPR and observe their effects on the physiological and
biochemical properties of rice plants grown under non-saline and saline glasshouse conditions.
Three strains were selected based on their salt-tolerance and plant growth-promoting properties under
in vitro saline conditions. These strains were identified as Bacillus tequilensis (UPMRB9), Bacillus
aryabhattai (UPMRE6), and Providencia stuartii (UPMRG1) using a 16S rRNA technique. The selected
strains were inoculated to three different rice varieties, namely BRRI dhan67 (salt-tolerant), Putra-1
(moderate salt-tolerant), and MR297 (salt-susceptible) under glasshouse conditions. Results showed
that the MR297 rice variety inoculated with UPMRB9 produced the highest total chlorophyll content,
with an increment of 28%, and the lowest electrolyte leakage of 92%. The Putra-1 rice variety also showed
a 156% total dry matter increase with the inoculation of this bacterial strain. The highest increase of
relative water content and reduction of Na/K ratio were found upon inoculation of UPMRE6 and
UPMRB9, respectively. The biggest significant effects of these bacterial inoculations were on relative
water content, electrolyte leakage, and the Na/K ratio of the BRRI dhan67 rice variety under saline
conditions, suggesting a synergistic effect on the mechanisms of plant salt-tolerance. This study
has shown that the application of locally-isolated salt-tolerant PGPR strains could be an effective
long-term and sustainable solution for rice cultivation in the coastal areas, which are affected by
global climate change.
- PGPR
- salt-tolerant
- rice
- salinity
- climate change
1. Definition
Salt-tolerant Plant Growth-Promoting Rhizobacteria (PGPR) has been reported to possess various salt-tolerant mechanisms, namely through the production of exopolysaccharide (EPS) and the formation of a biofilm, which have been proven to restrict Na+ uptake under saline soil conditions.
2. Introduction
Globally, plant growth and productivity are being hampered by various biotic and abiotic stresses [1]. Among them, salinity is regarded as one of the important agricultural problems, as it affects more than 20% of the total and irrigated land [2]. Rice (Oryza sativa L.) is one of the staple food crops across the globe and the demand is rising to feed the ever-increasing human population. Rice is a glycophyte plant by nature; it is inherently prone to salinity and will show a wide and vivid detrimental response to an increased salt level in the soil [3]. Moreover, the utilization of saline soil for agriculture has become necessary due to the lack of arable land. Several physical and chemical strategies for salinity mitigation have been tested but these methods are not feasible, causing adverse impacts on the ecosystem, thereby creating other problems. Therefore, identifying and developing an eco-friendly strategy that can ameliorate plant growth in response to abiotic stresses is vital in the current agricultural systems, which have to cope with the effect of climate change [4].
Having this in mind, the genetic technology to develop salt-tolerant rice variety and the utilization of PGPR could be the answer to improve rice cultivation on saline soils [5]. Several reports have been published on the effectiveness of PGPR on improving the growth of crops under environmental stress conditions [6][7]. In addition to the direct beneficial effect of PGPR, it has also been reported to conserve soil fertility, thus ensuring a favorable alternative to inorganic fertilizers and pesticides for sustainable agriculture systems [8]. Several bacterial genera, such as Alcaligenes, Azospirillum, Bacillus, Clostridium, Klebsiella, Pseudomonas, Rhizobium, Thiobacillus, Serratia, and Streptomyces, were used and tested as plant growth-promoters (PGP) under saline conditions [9]. Various researches have shown the commanding role of PGPRs in mitigating salt stress in different crop plants [10][11][12][13][14].
Plant growth-promoting rhizobacteria can play an important role during the seedling growth stage through several mechanisms, including biological N fixation, P solubilization, boosting of photosynthetic activities, chelation of iron through siderophores production, and synthesis of plant growth regulators such as indole-3-acetic acid (IAA) [15][16][17][18][19][20]. These positive traits of PGPR during the establishment of the crop seedling will significantly enhance plant growth during the later growth stage.
Bacterial flocculation or aggregate formation as a response to salinity, which are closely associated with the production of bacterial exopolysaccharides, help bacteria to survive in stressed environments and assist the host plant in tolerating various stresses [21]. Exopolysaccharides play a fundamental role in the formation of a bacterial biofilm, which enhances bacterial colonization on plant root surfaces [22]. Gholami et al. [23] showed enhanced germination of seed and vigorous growth of maize seedling by PGPR inoculation during the early plant growth stage, consequently resulting in an increased yield. According to Yoshida [24], the accumulation of essential nutrients for plant growth, such as N, P, K, and S, at the early stage of rice seedling growth is very crucial for the subsequent vegetative and reproductive stages.
The relationship between PGPR and plants has been widely known; however, there is a dearth of information regarding the effect of salt-tolerant PGPR on the growth of high yielding salt-responsive rice varieties. This is of utmost importance considering the perspective of salinity mitigation in coastal salt-affected rice-growing areas, in which seawater intrusion has become an urgent threat in recent years due to global climate change.
23. Data, Model, Applications and Influences
23.1. Salt-Tolerance and Plant Growth-Promoting Properties of Selected Bacterial Isolates
UPMRB9 produced the highest amount of exopolysaccharide (EPS) at 1.5 M of NaCl (30.50 g L−1), which is significantly higher than UPMRG1, UPMRE3, and UPMRA4 (Table 1 and Scheme 1). This particular strain also produced significantly higher biofilm and Na uptake compared to other strains. Meanwhile, UPMRE6 recorded the highest floc yield production (19.67 g L−1).
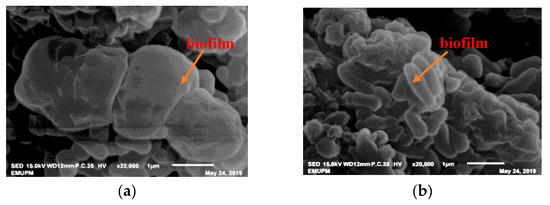
Scheme 1. Biofilm protects bacterial cells within EPS matrix of UPMRB9 (a) and UPMRE6 (b) at 1.5 M of NaCl concentration.
Table 1. Salt-tolerance and plant growth-promoting traits of selected bacterial isolates at 1.5 M of NaCl concentration.
| Bacterial Isolates | Salt-Tolerance Characteristics | Plant Growth-Promoting Characteristics | ||||||
| EPS (g L−1) | Floc Yield (g L−1) | Biofilm (590nm) | Sodium Uptake (mg L−1) | IAA (µg mL−1) | P Solubilization (µg mL−1) | K Solubilization (mg L−1) | Nitrogen Fixation | |
| UPMRA4 | 7.36c | 17.83a | 0.44d | 7.55cd | 6.71b | 9.21b | 2.08ab | − |
| UPMRB9 | 30.50a | 19.28a | 1.14a | 23.8a | 8.25a | 15.18a | 2.17ab | + |
| UPMRE3 | 12.24c | 9.73c | 0.31e | 6.40d | 3.50c | 9.34b | 0.70c | − |
| UPMRE6 | 26.18ab | 19.67a | 0.95b | 12.49bc | 6.70b | 15.30a | 2.97a | + |
| UPMRG1 | 21.79b | 12.90b | 0.77c | 13.51b | 6.99ab | 9.24b | 1.72bc | + |
|
Note: ‘+’ indicates positive, ‘−’ indicates negative. Means with the same letter in a column do not differ significantly using Tukey’s test at p > 0.05. EPS: exopolysaccharide; IAA: indole-3-acetic acid. |
||||||||
The plant growth-promoting characters showed that the UPMRB9 isolate produced significantly higher IAA (8.25 µg mL−1) compared to UPMRA4, UPMRE3, and UPMRE6. The latter strain had the highest P (15.30 µg mL−1) and K (2.97 mg L−1) solubilizers. The qualitative test showed that UPMRB9, UPMRE6, and UPMRG1 were N2 fixers.
23.2. Identification of PGPR Isolates Based on Partial 16S rRNA Gene Sequences
The PGPR isolates UPMRB9, UPMRE6, and UPMRG1 were selected for molecular identification based on their salt-tolerance and plant growth-promoting properties. The 16S rRNA fragments were successfully amplified using PCR. Approximately 1413 bp for UPMRB9 and UPMRG1 and 1411 bp for UPMRE6 were sequenced. The BLASTX comparison searches against the NCBI nucleotide database revealed 99% similarity of the isolates UPMRB9 with Bacillus tequilensis 10b (NCBI accession number: NR 104919.1), UPMRE6 with Bacillus aryabhattai B8W22 (NCBI accession number: NR 115953.1), and UPMRG1 with Providencia stuartii strain ATCC 29914 (NCBI accession number: NR 024848.1). The phylogenetic analyses of the PGPR isolates were[1] done based on neighbor-joining bootstrap analysis (Figure 1a,b).
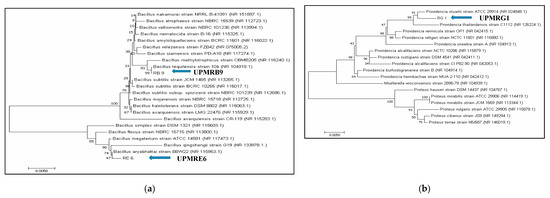
Figure 1. (a): Neighbor-joining tree based on the 16S rRNA gene sequence showing the phylogenetic relationship of UPMRB9 and UPMRE6 to related isolates; bar represents the nucleotide divergence value. (b): Neighbor-joining tree based on the 16S rRNA gene sequence showing the phylogenetic relationship of UPMRG1 to related isolates; bar represents the nucleotide divergence value.
23.3. Effect of Bacterial Inoculation on Total Chlorophyll Content of Three Rice Varieties
The total chlorophyll concentration of the rice plants was varied under non-saline and saline conditions. Under non-saline conditions, the total chlorophyll content of BRRI dhan67 treated with UPMRE6 increased by 19%, that of Putra-1 inoculated by UPMRB9 increased by 10% and that of MR297 treated with UPMRE6 increased by 15% (Figure 2a). On the contrary, under salt stress conditions, UPMRB9 inoculation produced the highest total chlorophyll content of all rice varieties with increments of 22%, 28%, and 28% for rice varieties BRRI dhan67, Putra-1, and MR297, respectively, compared to uninoculated plants (Figure 2b).
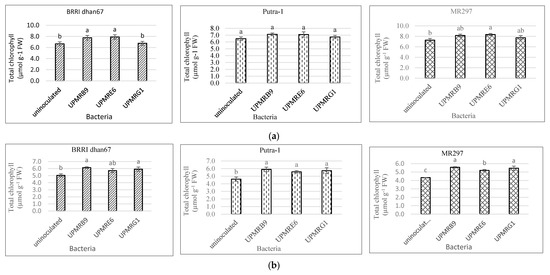
Figure 2. (a) Effect of bacterial inoculation on total chlorophyll content of three rice varieties under non-saline conditions. Means with the same letter in a column do not differ significantly using Tukey’s test at p > 0.05. (b) Effect of bacterial inoculation on total chlorophyll content of three rice varieties under saline conditions. Means with the same letter in a column do not differ significantly using Tukey’s test at p > 0.05.
23.4. Effect of Bacterial Inoculation on the Relative Water Content of Three Rice Varieties
Bacterial inoculation had a significant effect on an increasing percentage of relative water content both under non-saline and saline conditions (Figure 3a,b). Under non-saline conditions, the inoculation of UPMRE6 increased the relative water content of BRRI dhan67 and MR297 by 28% and 34%, respectively, while UPMRB9 increased the relative water content of Putra-1 by 28%. Under saline conditions, the relative water content of BRRI dhan67 and Putra-1 inoculated with UPMRB9 increased by 39% and 49%, respectively, while the relative water content of MR297 inoculated with UPMRE6 increased by 62%.
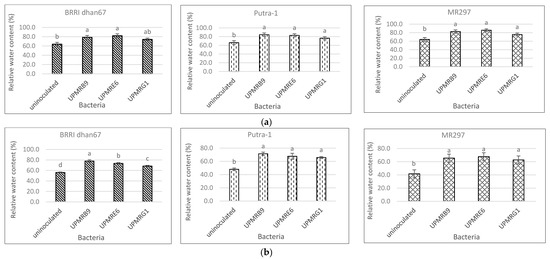
Figure 3. (a) Effect of bacterial inoculation on the relative water content of three rice varieties under non-saline conditions. Means with the same letter in a column do not differ significantly using Tukey’s test at p > 0.05. (b) Effect of bacterial inoculation on the relative water content of three rice varieties under saline conditions. Means with the same letter in a column do not differ significantly using Tukey’s test at p > 0.05.
23.5. Effect of Bacterial Inoculation on Electrolyte Leakage of Three Rice Varieties
Under non-saline conditions, electrolyte leakage of three rice varieties was not significantly varied by bacterial inoculation (data not shown), whilst under saline conditions, these values were significant across all inoculations (Figure 4). Leaf electrolyte leakage was significantly reduced with bacterial inoculation as compared to uninoculated plants. The highest reductions of electrolyte leakage in BRRI dhan67 and MR297 treated with UPMRB9 were 61% and 92%, respectively, while Putra-1 treated with UPMRG1 reduced the leakage by 55%.
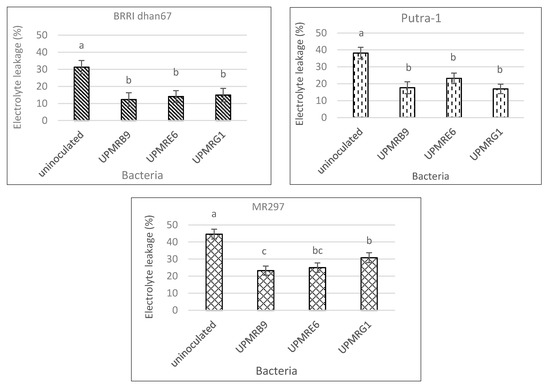
Figure 4. Effect of bacterial inoculation on electrolyte leakage of three rice varieties under saline conditions. Means with the same letter in a column do not differ significantly using Tukey’s test at p > 0.05.
23.6. Effect of Bacterial Inoculations on Total Dry Matter Production of Three Rice Varieties
Total dry matter (TDM) of the three rice varieties varied significantly due to the bacterial inoculations under both the non-saline and saline conditions. Bacterial inoculation increased TDM under non-saline conditions as compared to the uninoculated control, except for the inoculation of UPMRG1 on MR297 (Figure 5a).
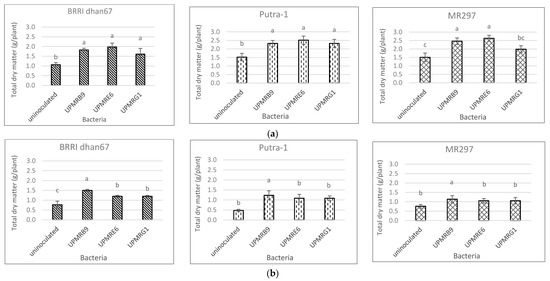
Figure 5. (a) Effect of bacterial inoculation on total dry matter production of three rice varieties under non-saline conditions. Means with the same letter in each variety do not differ significantly using Tukey’s test at p > 0.05. (b) Effect of bacterial inoculation on total dry matter production of three rice varieties under saline conditions. Means with the same letter in each variety do not differ significantly using Tukey’s test at p > 0.05.
Under saline conditions, UPMRB9 significantly produced higher TDM compared to other treatments for the BRRI dhan67, Putra-1, and MR297 varieties with increments of 56%, 156%, and 50%, respectively, compared to the uninoculated control (Figure 5b).
23.7. Effect of Salinity, PGPR and Rice Varieties on Na/K Ratio in the Above-Ground (Shoot) and Below Ground (Root and Soil) Parts of Three Rice Varieties
Under non-stress conditions, the Na/K ratio was not varied significantly (data not shown). Under saline conditions, the BRRI dhan67 rice plant treated with UPMRB9 had the greatest reduction, of 81%, in Na/K in its above-ground parts (Table 2).
Table 2. Effect of bacterial inoculation on the ratio of Na/K (% decrease) in the above-ground parts (shoots) of three rice varieties under saline soil conditions.
| Variety | Shoot | ||||||
| Uninoculated | UPMRB9 | UPMRE6 | UPMRG1 | ||||
| BRRI dhan67 | 0.73a | 0.14c (81%) | 0.22b (70%) | 0.26b (64%) | |||
| 0.81a (62%) | 0.67ab (34%) | 0.63b (26%) | Putra-1 | 1.13a | 0.39c (66%) | 0.57b (50%) | 0.60b (47%) |
| MR297 | 1.37a | 0.81c (41%) | 1.04b (24%) | 1.09b (20%) | |||
|
Means having the same letter in each variety do not differ significantly using Tukey’s test at p > 0.05. |
|||||||
A similar trend was observed for the Na/K ratio in the roots of BRRI dhan67 treated with UPMRB9, with a 53%reduction (Table 3). Interestingly, the soil sample showed the highest increment of the Na/K ratio in the MR297 rice plants treated with UPMRB9 (119%).
Table 3. Effect of bacterial inoculation on the Na/K ratio (% decrease/increase) in the below-ground parts (roots and soil) of three rice varieties under saline soil.
| Variety | Root | Soil | ||||||
| CONTROL | UPMRB9 | UPMRE6 | UPMRG1 | CONTROL | UPMRB9 | UPMRE6 | UPMRG1 | |
| BRRI dhan67 | 0.61a | 0.29c (53%) | 0.50b (18%) | 0.53b (13%) | 0.50b | |||
| Putra-1 | 0.79a | 0.90a (14%) | 0.42b (47%) | 0.45b (43%) | 0.46b | 0.64a (39%) | 0.48b (4%) | 0.46b (0%) |
| MR297 | 1.11a | 1.04a (6%) | 0.60b (46%) | 0.56b (50%) | 0.27b | 0.59a (119%) | 0.30b (11%) | 0.37b (37%) |
|
Means having the same letter in each variety do not differ significantly using Tukey’s test at p > 0.05. |
||||||||
23.8. SEM and TEM Observations of Bacterial Root Colonization
Electron micrograph observations revealed that the rice roots of BRRI dhan67 were successfully colonized with the inoculum under salt stress conditions. SEM showed that the UPMRB9 isolates were abundant around roots’ rhizosphere. The bacterial presence was observed (a) in between cell walls and the root hair zone and (b) in aggregate formation near root crevices (Scheme 2). The transverse sections of the root observed under TEM showed that UPMRB9 also inhabit in (a) the intercellular spaces within the epidermis and (b) near the vascular bundle along the cell wall within the epidermis (Scheme 3).

Scheme 2. SEM micrograph showing the colonization of UPMRB9 on roots of a BRRI dhan67 plant. The blue arrow shows UPMRB9 (a) in between cell walls and (b) near the crevices under saline conditions.
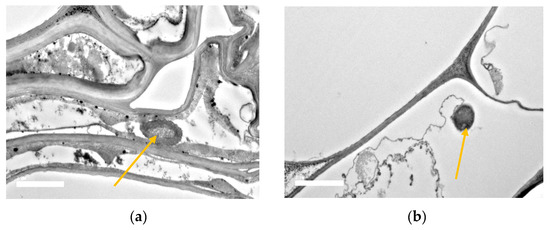
Scheme 3. TEM micrograph showing the colonization of UPMRB9 on roots of BRRI dhan67. The yellow arrow shows UPMRB9 (a) in intercellular spaces and (b) within the epidermis under saline conditions.
References
- I. Nemati; Foad Moradi; S. Gholizadeh; M.A. Esmaeili; M.R. Bihamta; The effect of salinity stress on ions and soluble sugars distribution in leaves, leaf sheaths and roots of rice (Oryza sativa L.) seedlings. Plant, Soil and Environment 2011, 57, 26-33, 10.17221/71/2010-pse.
- Samant A, Jawali N; Early seedling stage salt tolerance evaluation of genetically diverse rice genotypes. Ann. Biol. Res. 2016, 7, 46-54.
- Bhaswati Ghosh; Nasim Ali; Response of Rice under Salinity Stress: A Review Update. Rice Research: Open Access 2016, 4, 1-8, 10.4172/2375-4338.1000167.
- S Viscardi; V Ventorino; P Duran; A Maggio; Stefania De Pascale; M.L Mora; O Pepe; Assessment of plant growth promoting activities and abiotic stress tolerance of Azotobacter chroococcum strains for a potential use in sustainable agriculture. Journal of Soil Science and Plant Nutrition 2016, 16, 848-863, 10.4067/S0718-95162016005000060.
- Bhavanath Jha; Iti Gontia; Anton Hartmann; The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant and Soil 2011, 356, 265-277, 10.1007/s11104-011-0877-9.
- Salme Timmusk; Seong-Bin Kim; Eviatar Nevo; Islam Abd El Daim; Bo Ek; Jonas Bergquist; Lawrence Behers; Sfp-type PPTase inactivation promotes bacterial biofilm formation and ability to enhance wheat drought tolerance. Frontiers in Microbiology 2015, 6, 387, 10.3389/fmicb.2015.00387.
- Mahwish Zahid; M. Kaleem Abbasi; Sohail Hameed; Nasir Rahim; Isolation and identification of indigenous plant growth promoting rhizobacteria from Himalayan region of Kashmir and their effect on improving growth and nutrient contents of maize (Zea mays L.). Frontiers in Cellular and Infection Microbiology 2015, 6, 207, 10.3389/fmicb.2015.00207.
- Afshan Majeed; M. Kaleem Abbasi; Sohail Hameed; Asma Imran; Nasir Rahim; Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Frontiers in Cellular and Infection Microbiology 2015, 6, 198, 10.3389/fmicb.2015.00198.
- John M. Whipps; Microbial interactions and biocontrol in the rhizosphere. Journal Of Experimental Botany 2001, 52, 487-511, 10.1093/jexbot/52.suppl_1.487.
- Shimon Mayak; Tsipora Tirosh; Bernard R Glick; Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiology and Biochemistry 2004, 42, 565-572, 10.1016/j.plaphy.2004.05.009.
- D. Saravanakumar; R. Samiyappan; ACC deaminase from Pseudomonas fluorescens mediated saline resistance in groundnut (Arachis hypogea) plants. Journal of Applied Microbiology 2007, 102, 1283-1292, 10.1111/j.1365-2672.2006.03179.x.
- Pushp Sheel Shukla; Pradeep K. Agarwal; Bhavanath Jha; Improved Salinity Tolerance of Arachis hypogaea (L.) by the Interaction of Halotolerant Plant-Growth-Promoting Rhizobacteria. Journal of Plant Growth Regulation 2011, 31, 195-206, 10.1007/s00344-011-9231-y.
- Himadri Bhusan Bal; Lipika Nayak; Subhasis Das; Tapan K. Adhya; Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant and Soil 2012, 366, 93-105, 10.1007/s11104-012-1402-5.
- S. K. Upadhyay; D. P. Singh; Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biology 2014, 17, 288-293, 10.1111/plb.12173.
- Han HS, Lee KD; Plant growth-promoting rhizobacteria effect on antioxidant status, photosynthesis, mineral uptake and growth of lettuce under soil salinity. Res. J. Agric. Biol. Sci. 2005, 1, 210-215.
- M. Rajkumar; R. Nagendran; Kui Jae Lee; Wang Hyu Lee; Sung Zoo Kim; Influence of plant growth promoting bacteria and Cr6+ on the growth of Indian mustard. Chemosphere 2006, 62, 741-748, 10.1016/j.chemosphere.2005.04.117.
- Mohammad Saghir Khan; Almas Zaidi; Parvaze A. Wani; Role of phosphate-solubilizing microorganisms in sustainable agriculture — A review. Agronomy for Sustainable Development 2007, 27, 29-43, 10.1051/agro:2006011.
- Haiming Liu; Yanjing He; Haixia Jiang; Huasong Peng; Xian-Qing Huang; Xuehong Zhang; Linda S. Thomashow; Yuquan Xu; Characterization of a Phenazine-Producing Strain Pseudomonas chlororaphis GP72 with Broad-Spectrum Antifungal Activity from Green Pepper Rhizosphere. Current Microbiology 2007, 54, 302-306, 10.1007/s00284-006-0444-4.
- D. Saravanakumar; Charles Vijayakumar; N. Kumar; R. Samiyappan; PGPR-induced defense responses in the tea plant against blister blight disease. Crop Protection 2007, 26, 556-565, 10.1016/j.cropro.2006.05.007.
- Parvaze Ahmad Wani; Mohammad Saghir Khan; Almas Zaidi; Effect of metal tolerant plant growth promoting Bradyrhizobium sp. (vigna) on growth, symbiosis, seed yield and metal uptake by greengram plants. Chemosphere 2007, 70, 36-45, 10.1016/j.chemosphere.2007.07.028.
- Aisha Waheed Qurashi; Anjum Nasim Sabri; Biofilm formation in moderately halophilic bacteria is influenced by varying salinity levels. Journal of Basic Microbiology 2011, 52, 566-572, 10.1002/jobm.201100253.
- Yun Chen; Fang Yan; Yunrong Chai; Hongxia Liu; Roberto Kolter; Richard Losick; Jianhua Guo; Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation.. Environmental Microbiology 2012, 15, 848-864, 10.1111/j.1462-2920.2012.02860.x.
- Gholami A, Shahsavani S, Nezarat S; The effect of plant growth promoting rhizobacteria on seed germination and seedling growth of maize. Int. J. Biol. Sci. 2009, 1, 35-40.
- Yoshida S.. Fundamentals of Rice Crop Production; IRRI:: Los Banos, Philippines, 1981; pp. 1.
