The COP9 signalosome (CSN) is a regulator of the ubiquitin proteasome system (UPS). In mammalian cells it occurs as an eight-subunit protein complex, six Proteasome-COP9 signalosome-Initiation factor eIF3 (PCI)-domain subunits including CSN1-4, CSN7 and CSN8 and two MOV34-Pad1-N-terminal (MPN)-domain subunits called CSN5 and CSN6. The CSN regulates cullin-RING-ubiquitin ligases (CRLs) by specifically removing NEDD8 from cullins. In cooperation with CAND1 it controls the adaptation of the CRL network to fluctuations in substrate availability. The CSN complex belongs to the JAMM family of deubiquitylating enzymes (DUBs). In addition, it interacts with other deubiquitylating enzymes including USP15 and USP48 coordinating ubiquitylation and deubiquitylation activities.
- COP9 signalosome
- deubiquitylating enzymes
- ubiquitin
- deneddylation
1. Definition
The COP9 signalosome (CSN) is a signaling platform controlling the cellular ubiquitylation status. It determines the activity and remodeling of ~700 cullin-RING ubiquitin ligases (CRLs), which control more than 20% of all ubiquitylation events in cells and thereby influence virtually any cellular pathway. In addition, it is associated with deubiquitylating enzymes (DUBs) protecting CRLs from autoubiquitylation and rescuing ubiquitylated proteins from degradation. The coordination of ubiquitylation and deubiquitylation by the CSN is presumably important for fine-tuning the precise formation of defined ubiquitin chains.
2. Introduction
The COP9 signalosome (CSN) is a multiprotein complex representing a hallmark of eukaryotic cells. The CSN was discovered as a repressor of constitutive photomorphogenesis (COP) in Arabidopsis [1][2] and first isolated from cauliflower [3][4]. Purification from mammalian cells characterized the complex as signaling particle (signalosome) possessing homology to the 26S proteasome lid [5][6][7]. In Mammalia, core CSN is composed of six proteasome-COP9-initiation factor 3 (PCI) and two Mov34-and-Pad1p N-terminal (MPN) domain subunits [8][9], essential for CSN function. The 3.8 Å resolution CSN crystal structure based on human recombinant subunits provided detailed information about the subunit-subunit interactions [10]. The PCI domain proteins oligomerize via their winged-helix subdomains in the order of CSN7-CSN4-CSN2-CSN1-CSN3-CSN8, forming a horseshoe-like structure (Figure 1). The MPN domain heterodimer (CSN5, CSN6) is situated on top of the helical bundle formed by the C-terminal α-helices of each CSN subunit [10]. This architecture is shared by paralog complexes of the CSN: the 26S proteasome lid and the translation initiation factor 3 (eIF3) [10][11].
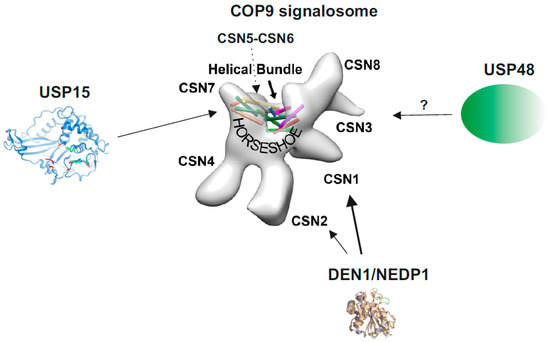
Figure 1. The COP9 signalosome (CSN) and its associated deubiquitylating enzymes (DUBs) and deneddylating enzyme 1 (DEN1). The structure of the CSN was obtained by cryo-electron microscopy using native CSN purified from human red blood cells or from mouse B8 fibroblasts [12]. The localization of CSN subunits, the “Helical Bundle” and the “Horseshoe” structure [10] is indicated. The crystal structure of USP15 is shown with its catalytic core (green) [13]. CSN5 is involved in CSN-USP15/Ubp12 interaction [14]. In addition, in A. nidulans USP15/UspA is presumably associated with the entire helical bundle [15]. So far, there is no crystal or cryo-structure of USP48 available. The crystal structure of DEN1/NEDP1 is from Shen et al. [16]. In human cells DEN1 mostly interacts with the N-terminus of CSN1, whereas in fungi it preferentially binds to CSN7 [17].
The CSN is substantially more heterogeneous than indicated by the structure of the eight-core subunits. It is associated with kinases and modified by phosphorylation [18][19][20]. For instance, in response to DNA damage causing double strand breaks the ATM kinase phosphorylates CSN3 [21] and UV damage leads to modification of CSN1 [22] with consequences for DNA damage repair. Thus, phosphorylation/dephosphorylation in response to signaling processes produces a dynamic heterogeneity of CSN complexes. Moreover, it has recently been recognized that a fraction of cellular CSN contains a non-essential, non-canonical component called CSNAP [23][24][25]. A further unexplored source of heterogeneity is provided by the fact that several CSN core subunits occur as paralogs/isoforms [26][27]. CSN subunit isoforms originate from gene duplication [26] or from use of alternative translation start sites as shown for CSN8A and CSN8B [28]. They are integrated into distinct CSN variants, which coexist in cells. In Arabidopsis, the CSN variants CSNCSN5A and CSNCSN5B confer different physiological functions [2] and in human cells CSNCSN7A and CSNCSN7B have distinct roles in adipogenic differentiation [26][29].
3. The CSN Belongs to the Deubiquitylating Enzymes (DUBs)
The ~100 DUBs encoded by the human genome belong to the following families: the ubiquitin (Ub)-specific protease (USP), Ub-C-terminal hydrolase (UCH), JAB1/MPN+/MOV34 protease (JAMM), ovarian tumor protease (OTU), Josephin [30], the novel motif interacting with Ub-containing DUB (MINDY) [31] and the recently discovered ZUFSP/ZUP1 family [32]. DUBs are pivotal regulators of the Ub system involved in protein turnover, signaling, sorting and trafficking [33]. Whereas most DUBs are cysteine proteases, the CSN and its paralog complex lid are metalloproteases of the JAMM family with conserved His, Asp and Ser coordinating a catalytic Zn2+ [30]. CSN5 is the only CSN subunit possessing the JAMM motif. Of note, free CSN5 is inactive [12][34] similar to its paralog subunit RPN11 of the 26S proteasome lid [35]. Furthermore, within the CSN complex, CSN5 is in an auto-inhibited state, the Ins-1 conformation [10]. Active CSN5 specifically removes NEDD8 from isopeptide-bonds with conserved Lys residues of cullins. The deneddylating activity of the CSN is activated by its substrates, neddylated cullin-RING-Ub ligases (NEDD8-CRLs) [36], which change the conformation of the active site called induced fit [37]. Structural data provide evidence for coordinated domain changes of CSN2, CSN4, and CSN7 and the CSN5-CSN6 heterodimer induced by neddylated CRL4A converting CSN5 into its active conformation [37]. Obviously, the CSN complex is an essential platform for CSN5 to act as a DUB. Purified mammalian CSN is a DUB specific for NEDD8 and unable to cleave Ub-AMC [12]. Using polyubiquitylated CUL4A as substrate and mutated CSN5, deubiquitylating activity was detected in crude Flag-CSN pulldowns from HeLa cells demonstrating the existence of CSN associated DUBs. Since wildtype CSN5 exhibited different deubiquitylating activity as compared to mutant CSN5, it was assumed that CSN5 has a deubiquitylating activity on its own [38]. However, the data might just reflect an impact of CSN5 on associated DUBs.
Neddylation [39] and deneddylation constitute a regulatory cycle, in which deneddylation inactivates CRLs [37][40] and NEDD8 conjugation stimulates CRL activity by multiple mechanisms [41][42][43][44]. Furthermore, CSN-mediated deneddylation is a prerequisite for the exchange of hundreds of substrate receptors (SRs) including F-box and BTB-domain SRs [45][46][47] as part of a rapid adaptation to altered protein degradation requirements [48]. In this process the CSN cooperates with Cullin-Associated and Neddylation-Dissociated 1 (CAND1) to accelerate the exchange of SRs to optimize the CRL network in response to fluctuations in substrate availability [45][46][49][50][51][52][53][54].
Recently, a potent and specific inhibitor of CSN-mediated deneddylating activity has been discovered, which is called CSN5i-3 [55]. The compound blocks cullin deneddylation and traps CRLs in the neddylated state. CSN5i-3 affects the viability of many tumor cells and suppresses growth of human xenografts in mice [55]. This excellent tool stimulates current and future research on CSN mechanisms and tumor therapy.
4. The CSN and Its Paralog 26S Proteasome Lid Cooperate with Diverse DUBs
Analyses of the CSN isolated from different cells by chromatography [14], pulldowns [29], immunoprecipitation [58] as well as density gradient centrifugation [17] revealed its association with additional DUBs, such as USP15 and USP48 and presumably other DUBs as well as with DEN1/NEDP1/SENP8, a member of the SENP family (Figure 1). Thus, the CSN occurs as a multi-DUB complex. USP15 and USP48 belong to the USPs, the largest DUB family, with more than 50 members in Mammalia [30]. Both are characterized by one (USP48) or two (USP15) UBL domains and are involved in multiple unrelated biochemical pathways and cellular responses. USP15 activity has been associated with parkin-mediated mitochondrial ubiquitylation and mitophagy [59] and the nuclear factor erythroid 2-related factor 2 pathway in an anti-oxidant response [60]. USP15 has also been shown to stabilize the CRL component RBX1 [61] as well as adenomatous polyposis coli (APC), a subunit of the β-catenin destruction complex [62] and to regulate transforming growth factor-β signaling [63]. There are less reports on USP48 engagements. USP48 stabilizes TRAF2 influencing the E-cadherin-mediated adherens junctions [64]. Whether all these USP15 and USP48 activities need CSN association is not clear at the moment. In the review we focus on CSN associated USP15 and USP48 and their functions in the NF-κB pathway as well as on DEN1, an associated deneddylase.
RPN11, the paralog to CSN5, is the intrinsic DUB of the lid, which also belongs to the JAMM-DUB family. Similar to CSN5, the Ins-1 loop of RPN11 undergoes conformational transition from inactive to active state, which is, in case of RPN11, directed by Ub and ATP [65]. In analogy to the CSN5-CSN6 heterodimer, RPN11 partners with another MPN domain protein, RPN8, possessing an inactive JAMM domain. The activated lid specifically cleaves Ub chains and promotes protein degradation by the 26S proteasome. A deneddylating activity of the lid was not reported. In the 19S regulatory particle the lid cooperates with USP14, a DUB of the USP family, and UCH37 of the UCH family. USP14 and UCH37 are not integral subunits of the 26S proteasome, they assist the lid in removing ubiquitin from substrates to ensure the function of the proteasome [66]. Interestingly, USP14 and UCH37 bind to RPN1 and to RPN13, respectively, which are, in addition to the RPN10, substrate receptors of the 19S regulatory particle [67], which provide a versatile binding platform for various ubiquitin chains [68]. Thus, a coordinated deubiquitylation of incoming substrates within the 19S regulator is presumably necessary for proper function of the 26S proteasome, which is accomplished by the cooperation of lid, USP14 and UCH37. However, since the lid is not directly associated with USP14 and UCH37, it is just part of a multi-DUB complex within the 19S particle.
Unfortunately, so far just few data are published on a possible intrinsic DUB activity of the other paralog complex, the eIF3 [69] and nothing is known about associated DUBs.
Similar structural principles as in the CSN and the lid are mirrored in the BRCA1-A complex in which the active JAMM domain DUB, BRCC36, interacts with the inactive JAMM protein ABRAXAS, whereas in the BRISC complex BRCC36 is supported by ABRO1 [70]. The main substrates of both complexes are Lys63-chains. Their functions, however, are completely different. Whereas BRCA1-A complex serves in DNA double-strand break repair sequestering BRCA1, the BRISC complex is involved in immune signaling. Thus, in this case complexes confer different targeting and specific regulatory functions to BRCC36 DUB, although the substrate remains the same [70]. In case of CSN and lid, the context of their multi-protein complexes provides substrate specificity to the DUBs as well as specific functions.
References
- Wei, N.; Deng, X.W. Cop9: A new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant Cell 1992, 4, 1507–1518.
- Qin, N.; Xu, D.; Li, J.; Deng, X.W. Cop9 signalosome: Discovery, conservation, activity, and function. J. Integr. Plant Biol. 2020, 62, 90–103.
- Wei, N.; Chamovitz, D.A.; Deng, X.W. Arabidopsis cop9 is a component of a novel signaling complex mediating light control of development. Cell 1994, 78, 117–124.
- Chamovitz, D.A.; Wei, N.; Osterlund, M.T.; von Arnim, A.G.; Staub, J.M.; Matsui, M.; Deng, X.W. The cop9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 1996, 86, 115–121.
- Seeger, M.; Kraft, R.; Ferrell, K.; Bech-Otschir, D.; Dumdey, R.; Schade, R.; Gordon, C.; Naumann, M.; Dubiel, W. A novel protein complex involved in signal transduction possessing similarities to 26s proteasome subunits. FASEB J. 1998, 12, 469–478.
- Wei, N.; Deng, X.W. Characterization and purification of the mammalian cop9 complex, a conserved nuclear regulator initially identified as a repressor of photomorphogenesis in higher plants. Photochem. Photobiol. 1998, 68, 237–241.
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the cop9-signalosome and eif3. Cell 1998, 94, 615–623.
- Hofmann, K.; Bucher, P. The pci domain: A common theme in three multiprotein complexes. Trends Biochem. Sci. 1998, 23, 204–205.
- Deng, X.W.; Dubiel, W.; Wei, N.; Hofmann, K.; Mundt, K.; Colicelli, J.; Kato, J.; Naumann, M.; Segal, D.; Seeger, M.; et al. Unified nomenclature for the cop9 signalosome and its subunits: An essential regulator of development. Trends Genet. 2000, 16, 202–203.
- Lingaraju, G.M.; Bunker, R.D.; Cavadini, S.; Hess, D.; Hassiepen, U.; Renatus, M.; Fischer, E.S.; Thoma, N.H. Crystal structure of the human cop9 signalosome. Nature 2014, 512, 161–165.
- des Georges, A.; Dhote, V.; Kuhn, L.; Hellen, C.U.; Pestova, T.V.; Frank, J.; Hashem, Y. Structure of mammalian eif3 in the context of the 43s preinitiation complex. Nature 2015, 525, 491–495.
- Rockel, B.; Schmaler, T.; Huang, X.; Dubiel, W. Electron microscopy and in vitro deneddylation reveal similar architectures and biochemistry of isolated human and flag-mouse cop9 signalosome complexes. Biochem. Biophys. Res. Commun. 2014, 450, 991–997.
- Ward, S.J.; Gratton, H.E.; Indrayudha, P.; Michavila, C.; Mukhopadhyay, R.; Maurer, S.K.; Caulton, S.G.; Emsley, J.; Dreveny, I. The structure of the deubiquitinase usp15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. J. Biol. Chem. 2018, 293, 17362–17374.
- Zhou, C.; Wee, S.; Rhee, E.; Naumann, M.; Dubiel, W.; Wolf, D.A. Fission yeast cop9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme ubp12p. Mol. Cell 2003, 11, 927–938.
- Meister, C.; Thieme, K.G.; Thieme, S.; Kohler, A.M.; Schmitt, K.; Valerius, O.; Braus, G.H. Cop9 signalosome interaction with uspa/usp15 deubiquitinase controls vea-mediated fungal multicellular development. Biomolecules 2019, 9, 238.
- Shen, L.N.; Liu, H.; Dong, C.; Xirodimas, D.; Naismith, J.H.; Hay, R.T. Structural basis of nedd8 ubiquitin discrimination by the deneddylating enzyme nedp1. EMBO J. 2005, 24, 1341–1351.
- Christmann, M.; Schmaler, T.; Gordon, C.; Huang, X.; Bayram, O.; Schinke, J.; Stumpf, S.; Dubiel, W.; Braus, G.H. Control of multicellular development by the physically interacting deneddylases den1/dena and cop9 signalosome. PLoS Genet. 2013, 9, e1003275.
- Uhle, S.; Medalia, O.; Waldron, R.; Dumdey, R.; Henklein, P.; Bech-Otschir, D.; Huang, X.; Berse, M.; Sperling, J.; Schade, R.; et al. Protein kinase ck2 and protein kinase d are associated with the cop9 signalosome. EMBO J. 2003, 22, 1302–1312.
- Bech-Otschir, D.; Kapelari, B.; Dubiel, W. The cop9 signalosome: Its possible role in the ubiquitin system. In Ubiquitin and the Chemistry of Life; Mayer, J.R., Ciechanover, A., Rechsteiner, M., Eds.; WILEY-VCH Verlag GmbH & KGaA: Weinheim, Germany, 2005; Volume 1, pp. 348–369.
- Fuzesi-Levi, M.G.; Ben-Nissan, G.; Bianchi, E.; Zhou, H.; Deery, M.J.; Lilley, K.S.; Levin, Y.; Sharon, M. Dynamic regulation of the cop9 signalosome in response to DNA damage. Mol. Cell. Biol. 2014, 34, 1066–1076.
- Meir, M.; Galanty, Y.; Kashani, L.; Blank, M.; Khosravi, R.; Fernandez-Avila, M.J.; Cruz-Garcia, A.; Star, A.; Shochot, L.; Thomas, Y.; et al. The cop9 signalosome is vital for timely repair of DNA double-strand breaks. Nucleic Acids Res. 2015, 43, 4517–4530.
- Dubois, E.L.; Gerber, S.; Kisselev, A.; Harel-Bellan, A.; Groisman, R. Uv-dependent phosphorylation of cop9/signalosome in uv-induced apoptosis. Oncol. Rep. 2016, 35, 3101–3105.
- Rozen, S.; Fuzesi-Levi, M.G.; Ben-Nissan, G.; Mizrachi, L.; Gabashvili, A.; Levin, Y.; Ben-Dor, S.; Eisenstein, M.; Sharon, M. Csnap is a stoichiometric subunit of the cop9 signalosome. Cell Rep. 2015, 13, 585–598.
- Fuzesi-Levi, M.G.; Fainer, I.; Ivanov Enchev, R.; Ben-Nissan, G.; Levin, Y.; Kupervaser, M.; Friedlander, G.; Salame, T.M.; Nevo, R.; Peter, M.; et al. Csnap, the smallest csn subunit, modulates proteostasis through cullin-ring ubiquitin ligases. Cell Death Differ. 2020, 27, 984–998.
- Gutierrez, C.; Chemmama, I.E.; Mao, H.; Yu, C.; Echeverria, I.; Block, S.A.; Rychnovsky, S.D.; Zheng, N.; Sali, A.; Huang, L. Structural dynamics of the human cop9 signalosome revealed by cross-linking mass spectrometry and integrative modeling. Proc. Natl. Acad. Sci. USA 2020, 117, 4088–4098.
- Dubiel, D.; Rockel, B.; Naumann, M.; Dubiel, W. Diversity of cop9 signalosome structures and functional consequences. FEBS Lett. 2015, 589, 2507–2513.
- Jin, D.; Li, B.; Deng, X.W.; Wei, N. Plant cop9 signalosome subunit 5, csn5. Plant Sci. 2014, 224C, 54–61.
- Lykke-Andersen, K.; Wei, N. Gene structure and embryonic expression of mouse cop9 signalosome subunit 8 (csn8). Gene 2003, 321, 65–72.
- Huang, X.; Ordemann, J.; Pratschke, J.; Dubiel, W. Overexpression of cop9 signalosome subunits, csn7a and csn7b, exerts different effects on adipogenic differentiation. FEBS Open Bio 2016, 6, 1102–1112.
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from genes to organism. Physiol. Rev. 2013, 93, 1289–1315.
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. Mindy-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 2016, 63, 146–155.
- Kwasna, D.; Abdul Rehman, S.A.; Natarajan, J.; Matthews, S.; Madden, R.; De Cesare, V.; Weidlich, S.; Virdee, S.; Ahel, I.; Gibbs-Seymour, I.; et al. Discovery and characterization of zufsp/zup1, a distinct deubiquitinase class important for genome stability. Mol. Cell 2018, 70, 150–164 e156.
- Clague, M.J.; Coulson, J.M.; Urbe, S. Cellular functions of the dubs. J. Cell Sci. 2012, 125, 277–286.
- Echalier, A.; Pan, Y.; Birol, M.; Tavernier, N.; Pintard, L.; Hoh, F.; Ebel, C.; Galophe, N.; Claret, F.X.; Dumas, C. Insights into the regulation of the human cop9 signalosome catalytic subunit, csn5/jab1. Proc. Natl. Acad. Sci. USA 2013, 110, 1273–1278.
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., 3rd; Koonin, E.V.; Deshaies, R.J. Role of rpn11 metalloprotease in deubiquitination and degradation by the 26s proteasome. Science 2002, 298, 611–615.
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of jab1/csn5 in cleavage of nedd8 from cul1. Science 2002, 298, 608–611.
- Cavadini, S.; Fischer, E.S.; Bunker, R.D.; Potenza, A.; Lingaraju, G.M.; Goldie, K.N.; Mohamed, W.I.; Faty, M.; Petzold, G.; Beckwith, R.E.; et al. Cullin-ring ubiquitin e3 ligase regulation by the cop9 signalosome. Nature 2016, 531, 598–603.
- Groisman, R.; Polanowska, J.; Kuraoka, I.; Sawada, J.; Saijo, M.; Drapkin, R.; Kisselev, A.F.; Tanaka, K.; Nakatani, Y. The ubiquitin ligase activity in the ddb2 and csa complexes is differentially regulated by the cop9 signalosome in response to DNA damage. Cell 2003, 113, 357–367.
- Liakopoulos, D.; Doenges, G.; Matuschewski, K.; Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998, 17, 2208–2214.
- Mosadeghi, R.; Reichermeier, K.M.; Winkler, M.; Schreiber, A.; Reitsma, J.M.; Zhang, Y.; Stengel, F.; Cao, J.; Kim, M.; Sweredoski, M.J.; et al. Structural and kinetic analysis of the cop9-signalosome activation and the cullin-ring ubiquitin ligase deneddylation cycle. Elife 2016, 5, e12102.
- Kawakami, T.; Chiba, T.; Suzuki, T.; Iwai, K.; Yamanaka, K.; Minato, N.; Suzuki, H.; Shimbara, N.; Hidaka, Y.; Osaka, F.; et al. Nedd8 recruits e2-ubiquitin to scf e3 ligase. EMBO J. 2001, 20, 4003–4012.
- Sakata, E.; Yamaguchi, Y.; Miyauchi, Y.; Iwai, K.; Chiba, T.; Saeki, Y.; Matsuda, N.; Tanaka, K.; Kato, K. Direct interactions between nedd8 and ubiquitin e2 conjugating enzymes upregulate cullin-based e3 ligase activity. Nat. Struct. Mol. Biol. 2007, 14, 167–168.
- Saha, A.; Deshaies, R.J. Multimodal activation of the ubiquitin ligase scf by nedd8 conjugation. Mol. Cell 2008, 32, 21–31.
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into nedd8 activation of cullin-ring ligases: Conformational control of conjugation. Cell 2008, 134, 995–1006.
- Dubiel, D.; Ordemann, J.; Pratschke, J.; Dubiel, W.; Naumann, M. Cand1 exchange factor promotes keap1 integration into cullin 3-ring ubiquitin ligase during adipogenesis. Int. J. Biochem. Cell Biol. 2015, 66, 95–100.
- Reitsma, J.M.; Liu, X.; Reichermeier, K.M.; Moradian, A.; Sweredoski, M.J.; Hess, S.; Deshaies, R.J. Composition and regulation of the cellular repertoire of scf ubiquitin ligases. Cell 2017, 171, 1326–1339.e14.
- Schmidt, M.W.; McQuary, P.R.; Wee, S.; Hofmann, K.; Wolf, D.A. F-box-directed crl complex assembly and regulation by the csn and cand1. Mol. Cell 2009, 35, 586–597.
- Dubiel, W.; Dubiel, D.; Wolf, D.A.; Naumann, M. Cullin 3-based ubiquitin ligases as master regulators of mammalian cell differentiation. Trends Biochem. Sci. 2018, 43, 95–107.
- Wu, S.; Zhu, W.; Nhan, T.; Toth, J.I.; Petroski, M.D.; Wolf, D.A. Cand1 controls in vivo dynamics of the cullin 1-ring ubiquitin ligase repertoire. Nat. Commun. 2013, 4, 1642.
- Zemla, A.; Thomas, Y.; Kedziora, S.; Knebel, A.; Wood, N.T.; Rabut, G.; Kurz, T. Csn- and cand1-dependent remodelling of the budding yeast scf complex. Nat. Commun. 2013, 4, 1641.
- Pierce, N.W.; Lee, J.E.; Liu, X.; Sweredoski, M.J.; Graham, R.L.; Larimore, E.A.; Rome, M.; Zheng, N.; Clurman, B.E.; Hess, S.; et al. Cand1 promotes assembly of new scf complexes through dynamic exchange of F box proteins. Cell 2013, 153, 206–215.
- Dubiel, D.; Gierisch, M.E.; Huang, X.; Dubiel, W.; Naumann, M. Cand1-dependent control of cullin 1-ring ub ligases is essential for adipogenesis. Biochim. Biophys. Acta 2013, 1833, 1078–1084.
- Liu, X.; Reitsma, J.M.; Mamrosh, J.L.; Zhang, Y.; Straube, R.; Deshaies, R.J. Cand1-mediated adaptive exchange mechanism enables variation in f-box protein expression. Mol. Cell 2018, 69, 773–786.e6.
- Straube, R.; Shah, M.; Flockerzi, D.; Wolf, D.A. Trade-off and flexibility in the dynamic regulation of the cullin-ring ubiquitin ligase repertoire. PLoS Comput. Biol. 2017, 13, e1005869.
- Schlierf, A.; Altmann, E.; Quancard, J.; Jefferson, A.B.; Assenberg, R.; Renatus, M.; Jones, M.; Hassiepen, U.; Schaefer, M.; Kiffe, M.; et al. Targeted inhibition of the cop9 signalosome for treatment of cancer. Nat. Commun. 2016, 7, 13166.
- Teixeira, L.K.; Reed, S.I. Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 2013, 82, 387–414.
- Chung, D.; Dellaire, G. The role of the cop9 signalosome and neddylation in DNA damage signaling and repair. Biomolecules 2015, 5, 2388–2416.
- Schweitzer, K.; Naumann, M. Csn-associated usp48 confers stability to nuclear nf-kappab/rela by trimming k48-linked ub-chains. Biochim. Biophys. Acta 2015, 1853, 453–469.
- Cornelissen, T.; Haddad, D.; Wauters, F.; Van Humbeeck, C.; Mandemakers, W.; Koentjoro, B.; Sue, C.; Gevaert, K.; De Strooper, B.; Verstreken, P.; et al. The deubiquitinase usp15 antagonizes parkin-mediated mitochondrial ubiquitination and mitophagy. Hum. Mol. Genet. 2014, 23, 5227–5242.
- Villeneuve, N.F.; Tian, W.; Wu, T.; Sun, Z.; Lau, A.; Chapman, E.; Fang, D.; Zhang, D.D. Usp15 negatively regulates nrf2 through deubiquitination of keap1. Mol. Cell 2013, 51, 68–79.
- Hetfeld, B.K.; Helfrich, A.; Kapelari, B.; Scheel, H.; Hofmann, K.; Guterman, A.; Glickman, M.; Schade, R.; Kloetzel, P.M.; Dubiel, W. The zinc finger of the csn-associated deubiquitinating enzyme usp15 is essential to rescue the e3 ligase rbx1. Curr. Biol. 2005, 15, 1217–1221.
- Huang, X.; Langelotz, C.; Hetfeld-Pechoc, B.K.; Schwenk, W.; Dubiel, W. The cop9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via usp15. J. Mol. Biol. 2009, 391, 691–702.
- Inui, M.; Manfrin, A.; Mamidi, A.; Martello, G.; Morsut, L.; Soligo, S.; Enzo, E.; Moro, S.; Polo, S.; Dupont, S.; et al. Usp15 is a deubiquitylating enzyme for receptor-activated smads. Nat. Cell Biol. 2011, 13, 1368–1375.
- Li, S.; Wang, D.; Zhao, J.; Weathington, N.M.; Shang, D.; Zhao, Y. The deubiquitinating enzyme usp48 stabilizes traf2 and reduces e-cadherin-mediated adherens junctions. FASEB J. 2018, 32, 230–242.
- Worden, E.J.; Dong, K.C.; Martin, A. An aaa motor-driven mechanical switch in rpn11 controls deubiquitination at the 26s proteasome. Mol. Cell 2017, 67, 799–811.e8.
- de Poot, S.A.H.; Tian, G.; Finley, D. Meddling with fate: The proteasomal deubiquitinating enzymes. J. Mol. Biol. 2017, 429, 3525–3545.
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124.
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19s cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 477.
- Moretti, J.; Chastagner, P.; Gastaldello, S.; Heuss, S.F.; Dirac, A.M.; Bernards, R.; Masucci, M.G.; Israel, A.; Brou, C. The translation initiation factor 3f (eif3f) exhibits a deubiquitinase activity regulating notch activation. PLoS Biol. 2010, 8, e1000545.
- Rabl, J.; Bunker, R.D.; Schenk, A.D.; Cavadini, S.; Gill, M.E.; Abdulrahman, W.; Andres-Pons, A.; Luijsterburg, M.S.; Ibrahim, A.F.M.; Branigan, E.; et al. Structural basis of brcc36 function in DNA repair and immune regulation. Mol. Cell 2019, 75, 483–497 e489.
