Centromeres are the complex structures responsible for the proper segregation of chromosomes during cell division. Structural or functional alterations of the centromere cause aneuploidies and other chromosomal aberrations that can induce cell death with consequences on health and survival of the organism as a whole. Because of their essential function in the cell, centromeres have evolved high flexibility and mechanisms of tolerance to preserve their function following stress, whether it is originating from within or outside the cell.Despite the differences in DNA sequences, protein composition and centromere size, all of these diverse centromere structures promote efficient chromosome segregation, balancing genome stability and adaptability, and ensuring faithful genome inheritance at each cellular generation.
- centromere
- neocentromere
- holocentromere
- CENP-A
- repetitive sequences
- centromere evolution
1.
Introduction
Centromeres are specialized chromatin regions that establish the assembly site for the kinetochore, a complex protein structure that mediates the attachment of spindle microtubules to chromosomes, thus permitting proper chromosome segregation during cell division. In all organisms studied thus far, it has been shown that no DNA sequence is either necessary or sufficient for centromere identity. The only known exception is Saccharomyces cerevisiae, whose centromeres are specified by a conserved 125-bp sequence (reviewed in [1]). Instead, centromeres are defined by the deposition of the histone H3 variant centromeric protein A (CENP-A) that replaces canonical histone H3 in centromeric nucleosomes [2][3][4][2–4]. CENP-A chromatin underlies the formation of the constitutive centromere-associated network (CCAN) [5][6][7][8][9][5–9], and in mitosis, serves as a template for assembly of the kinetochore to enable the chromosome for the correct segregation [10]. CENP-A is recruited at different stages of the cell cycle depending on the organism but, unlike canonical histones, its loading is uncoupled from DNA replication [11][12][11,12]. In human cells, CENP-A deposition occurs in late telophase or early G1 [13]. In Drosophila, Cid (homolog of CENP-A) is incorporated at different times depending on developmental stage and on the specific cellular culture, but it is generally also found to be loaded in late mitosis/early G1 [14][15][16][17][18][14–18]. In S. pombe on the other hand, CENP-A homolog is incorporated during G2 [19]. Although centromere DNA sequences are not conserved between species, and in some cases not even between centromeres of the same species, they generally contain DNA rich in repeated sequences, in particular tandem satellite DNA such as human alpha-satellite that can extend for mega bases, or SATIII as seen in Drosophila and in humans. Recent works have shown the centromeric presence of mobile elements, specifically retrotransposons, in several species including Drosophila, [20][21][20,21], humans [22] and maize [23], probably contribute to the establishment and maintenance of eukaryotic centromeres while promoting their variability (reviewed in [24][25][24,25]).
2. Centromere Flexibility in Response to Stress
Centromere Flexibility in Response to Stress
Because of their essential function in the cell, centromeres may have evolved high flexibility and mechanisms of tolerance to preserve their functionality following stress originating from within or outside the cell. Indeed, substantial changes in centromere integrity and overall size can cause chromosome aneuploidy, segregation and structural defects (reviewed in [26]) that can induce cell death with consequences on health and survival of the organism as a whole. DNA damage to the centromere may have multiple origins (reviewed in [26]). First of all, centromeres are subjected to mechanical stress during anaphase due to the microtubules that pull them towards the poles. Moreover, it has been proposed that alterations of the mitotic spindle are a possible cause of segregation and structural defects. In addition, spindle defects, that expose chromosomes to excessive forces, can generate centromeric double-strand breaks (DSBs), possibly leading to carcinogenesis [27]. Defects in DNA replication is another possible cause of stress for centromeres (reviewed in [28]). Because of their repetitive nature, the centromeric chromatin forms complex secondary structures [29][30][29,30], representing a problem during replication and inducing a stalled fork. This could make this region prone to replication errors and recombination events that disrupt the integrity or structure of the centromere, causing aneuploidy (reviewed in [31]). The centromere also responds to stimuli that reach the cell from the external environment. Any perturbing agent that changes the cellular microenvironment can be considered a source of stress and potentially harmful to the centromere’s essential function. Stressing factors can be both abiotic, such as heat, cold, UV light, heavy metals etc. [32][33][34][32–34], and biotic, such as parasites and infectious agents. Physiological changes derived from development and differentiation are also underlined by profound epigenetic and transcriptional transitions that contribute to diverse forms of stress for the cell [35]. Stressors that directly challenge the integrity of the genome by generating DNA damage or perturbing the DNA replication process can also impinge on centromeres. Notably, centromere DNA instability has also been associated with cancer and cellular senescence [36]. In the last years, several studies on different organisms have shown that heat shock induces transcriptional activation of centromeric and pericentromeric regions [37][38][39][40][41][37-41] (reviewed in [42][43][42,43]). Nevertheless, both centromeric and pericentromeric transcripts, with or without induction by external agents, have been implicated in various cellular functions, such as the transmission of epigenetic information, differentiation, and the cellular defense to stress [32][44][45][46][32,44,45,46] (reviewed in [42]). It is also widely assumed that transcription is a process closely related to the centromeric function in several organisms, including fission yeast [47][48][47,48], humans [49][50][49,50], and Drosophila [51][52][51,52], and that it is particularly associated with CENP-A deposition [53][54][55][56][53-56]. Indeed, it has been supposed that transcription is coupled to CENP-A loading and that it is required for CENP-A deposition into centromeric chromatin. In addition to repetitive satellite sequences, transposable elements (TEs) are abundant components of (peri)centromeric heterochromatin, as shown in humans [57][58][57,58] and Drosophila [59][60][59,60]. Centromeric TEs, in addition to satellite sequences, have also been shown to be transcribed [21][54][61][62][21,54,61,62] (reviewed in [63]). Some models have been proposed where retrotransposons could produce non-coding RNAs with a role in the centromere specification [22][64][22,64]. In Drosophila, but also in human cells, the analysis on immunostained chromatin fibers has shown a clear presence of H3K4me2 and H3K36me, typically associated with active chromatin in Cid (CENP-A in Drosophila) labeled centromeric chromatin, and of H3K9me3, a repressive histone marker in adjacent heterochromatin [65][66][65,66]. Recent studies have demonstrated that transcriptional activators of euchromatic genes belonging to the trx-G group, in particular Trithorax (Trx), Ash1 and CBP, co-localize with Cid-containing chromatin [67]. Ash1 and CBP depletion through post-transcriptional silencing of the respective coding genes causes a decrease in Cid at the centromere and a significant increase in chromosomal aberrations at all phases of mitosis, such as decondensation, lagging chromosomes and the generation of chromosomal fragments. Instead, Trx depletion causes the same chromosomal aberrations without affecting the overall level of Cid protein. Immunofluorescence analysis using antibodies against H3 histone has shown that Trx functions open up the chromatin, making it accessible to transcription factors. In fact, Trx depletion induces a compaction of the centromeric chromatin with a higher concentration of H3-containing nucleosomes. Ash1 and CBP are transcriptional activators which work through histone modifications. In particular, Ash1 methylates H3K4me2 and CBP acetylates H3K27ac at the centromere. Both modifications are specific for active chromatin and their decrease is related to a depletion of Cid [67]. It has been proposed that a balance between methylation and acetylation could create an epigenetic environment that favors Cid deposition [68]. Alternatively, it could be hypothesized that the activating epigenetic modifications have the function of preserving a euchromatic region inside the heterochromatic domain necessary for CENP-A/Cid loading (Figure 1). Whichever the mechanism, the failure to open the centromeric chromatin is incompatible with Cid deposition. It is not known whether the euchromatic epigenetic environment directly favors the Cid deposition through recruitment of Cal1 chaperone or whether it favors the transcription of centromeric sequences such as Sat III or centromeric transposons, which in turn are required for Cid deposition. Disentangling these two roles would be an important issue to address.
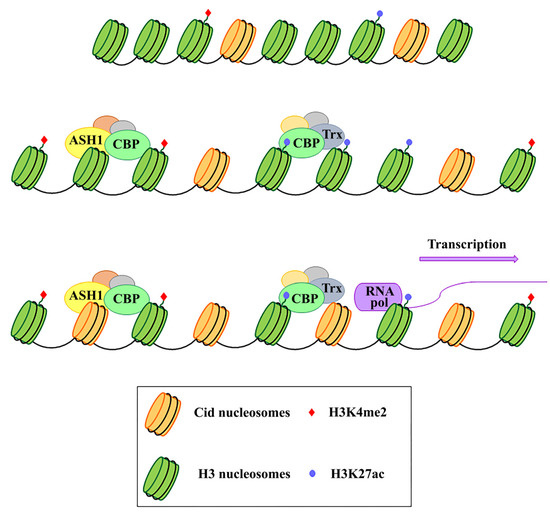
Figure 1.
It has been shown that Sat III in both Drosophila and mammals, and centromeric transposons, are weakly transcribed [21,32], but it is not clear whematic representation of theat their role in centromeric function could be. There is also the possibility that centromeres’ basal transcription is not causative but a mere consequence of the chromatin opening for Cid/CENP-A loading, without holding being necessary for centromeric functional role of Ash1, CBP, and Trx proteins in the centity. However, given several studies have shown that the direct inhibition of centromeric RNAs causes overall reduction in CENP-A at centromeres [50,51,54,69-74], the alternative possibility would imply that transcription may be a consequence of chromatin opening, while transcription products play a direct role in CENP-A deposition and centromeric stability. Despite the differences in DNA sequences, proteins composition and centromere size, all of these diverse centromere structures promote efficient chromosome segregation, balancing genome stability and adaptability, and ensuring faithful genome inheritance at each cellular generation.
Neocentromere as a Functional and Evolutionary Model for Centromere Biology
How and why neocentromeres appear icn a given chromosome region. These proteins work by are intriguing questions. It has been proposed that de novo centromeres might represent different scenarios: “latent” centromeres [75-77], which are locations of ancestral centromeres following centromere repositioning events [78], (reviewed in [79]), or euchromatic regions, where centromeric markers have spread from adjacent areas, inducing neocentromere formation near endogenous centromeres. Some studies in Drosophila support the model that formatifying the epigenetic state of the on of the neocentromere may depend on the proximity to an endogenous centromere. However, some observations suggest that neocentromeres are often formed in distal chromosomal regions, usually separated from endogenous centromeres by long tracts of DNA, and they cannot be explained as formed by the spread of centromeric cactivity in cis [80]. A different model for neocentromere formation called the “lateromatin. In particular, on H3 histones, Ash1 dimethylates lysine 4 (H3K4me2) and CBP acetylates lysine 27 (H3K27ac) within the al inhibition model” [81] supports the existence of many sites along a chromosome that are potentially able to perform centromeric activity, but that are normally repressed by a dominant centromere. It has been shown that a neocentromere is capable of forming near telomeres at the end of the chromosome [82]. This substantiates the hypothesis formulated by Agudo et al. [83] (reviewed in [84]) that the centromere evolved from telomere during the evolutionary origin of the eukaryotic chromosome. Recently, Palladino et al. [82] has shown that targeting Cal1 to chromosomal regions outside the original centromere induces the deposition of Cid and the formation of a new centromere at different locations, even at large genomic distance from the endogenous site. Zeitlin et al. [85] speculated that a neocentromere could emerge at site of breaks, following the observation that CENP-A is rapidly recruited to DNA double-strand breaks. This point of view is also supported by Ventura et al. [78], who noticed the closeness of the breakpoint to the neocentromere location in some studies. It has been shown that CENP-A is produced in excess of the needs for centromeres and that it is deposited around the original centromere and on islands of nucleosomes scattered along the chromosomes [21,86,87], only to be cleared during replication [21,88]. It is possible that some of these islands accumulate a quantity of CENP-A that predisposes them to acquire a potential centromeric function in case the endogenous centromeric region. Trx works by inducing chromatin opening which, in turn, favours bothe is damaged. In this case, not all chromosomal regions are equally and simultaneously predisposed to acquire a centromeric activity. Another mode of neocentromere formation may involve transposable elements (TE). Notably, stress induces the activation of TE, causing their transposition to new locations [41,89-92]. It is then possible that following chromosomal insult or rearrangements, a single TE may be sufficient to initiate CENP-A recruitment via its nascent transcription, breaks generation and other specific chromatin alterations associated with the “jump” and/or the TE reintegration into a new locus [64]. However, we favor an alternative hypothesis, that a random insertion of a TE per se does not trigger neocentromere formation unless it hits a CENP-A ectopic site. The concomitance of preexisting ectopic CENP-A and transposition may represent the first step towards the formation of a new centromere. Over time, the accumulation of other repetitive sequences through new transpositions or duplications of existing sequences in these CENP-A/Cid depos ectopic “hot spots” would create a genetic and epigenetic landscape for the evolution of a complex centromere that can functionally replace the endogenous one.
Holocentromere
Nematodes, some insection and activation of transcriptions and species of plants belonging to the flowering plants have diffused centromeres throughout their chromosomes, and these are described as “holocentric”. From a cytological point of view, holocentric chromosomes do not present a primary constriction in metaphase—a hallmark narrowing found in stereotypical metacentric or sub-metacentric chromosomes—and do not have a designated place along the chromosome for spindle microtubule attachment.
It has been shown that Sat III in both Drosophila and mammals, and centromeric transposons, are weakly transcribed [21][32], but it is not clear what their role in centromeric function could be. There is also the possibility that centromeres’ basal transcription is not causative but a mere consequence of the chromatin opening for Cid/CENP-A loading, without holding being necessary for centromeric functionality. However, given several studies have shown that the direct inhibition of centromeric RNAs causes overall reduction in CENP-A at centromeres [50][51][54][69][70][71][72][73][74], the alternative possibility would imply that transcription may be a consequence of chromatin opening, while transcription products play a direct role in CENP-A deposition and centromeric stability. Despite the differences in DNA sequences, proteins composition and centromere size, all of these diverse centromere structures promote efficient chromosome segregation, balancing genome stability and adaptability, and ensuring faithful genome inheritance at each cellular generation.
3. Neocentromere as a Functional and Evolutionary Model for Centromere Biology
How and why neocentromeres appear in a given chromosome region are intriguing questions. It has been proposed that de novo centromeres might represent different scenarios: “latent” centromeres [75][76][77], which are locations of ancestral centromeres following centromere repositioning events [78], (reviewed in [79]), or euchromatic regions, where centromeric markers have spread from adjacent areas, inducing neocentromere formation near endogenous centromeres. Some studies in Drosophila support the model that formation of the neocentromere may depend on the proximity to an endogenous centromere. However, some observations suggest that neocentromeres are often formed in distal chromosomal regions, usually separated from endogenous centromeres by long tracts of DNA, and they cannot be explained as formed by the spread of centromeric activity in cis [80]. A different model for neocentromere formation called the “lateral inhibition model” [81] supports the existence of many sites along a chromosome that are potentially able to perform centromeric activity, but that are normally repressed by a dominant centromere. It has been shown that a neocentromere is capable of forming near telomeres at the end of the chromosome [82]. This substantiates the hypothesis formulated by Agudo et al. [83] (reviewed in [84]) that the centromere evolved from telomere during the evolutionary origin of the eukaryotic chromosome. Recently, Palladino et al. [82] has shown that targeting Cal1 to chromosomal regions outside the original centromere induces the deposition of Cid and the formation of a new centromere at different locations, even at large genomic distance from the endogenous site. Zeitlin et al. [85] speculated that a neocentromere could emerge at site of breaks, following the observation that CENP-A is rapidly recruited to DNA double-strand breaks. This point of view is also supported by Ventura et al. [78], who noticed the closeness of the breakpoint to the neocentromere location in some studies. It has been shown that CENP-A is produced in excess of the needs for centromeres and that it is deposited around the original centromere and on islands of nucleosomes scattered along the chromosomes [21][86][87], only to be cleared during replication [21][88]. It is possible that some of these islands accumulate a quantity of CENP-A that predisposes them to acquire a potential centromeric function in case the endogenous centromere is damaged. In this case, not all chromosomal regions are equally and simultaneously predisposed to acquire a centromeric activity. Another mode of neocentromere formation may involve transposable elements (TE). Notably, stress induces the activation of TE, causing their transposition to new locations [41][89][90][91][92]. It is then possible that following chromosomal insult or rearrangements, a single TE may be sufficient to initiate CENP-A recruitment via its nascent transcription, breaks generation and other specific chromatin alterations associated with the “jump” and/or the TE reintegration into a new locus [64]. However, we favor an alternative hypothesis, that a random insertion of a TE per se does not trigger neocentromere formation unless it hits a CENP-A ectopic site. The concomitance of preexisting ectopic CENP-A and transposition may represent the first step towards the formation of a new centromere. Over time, the accumulation of other repetitive sequences through new transpositions or duplications of existing sequences in these CENP-A ectopic “hot spots” would create a genetic and epigenetic landscape for the evolution of a complex centromere that can functionally replace the endogenous one.
4. Holocentromere
Nematodes, some insects and species of plants belonging to the flowering plants have diffused centromeres throughout their chromosomes, and these are described as “holocentric”. From a cytological point of view, holocentric chromosomes do not present a primary constriction in metaphase—a hallmark narrowing found in stereotypical metacentric or sub-metacentric chromosomes—and do not have a designated place along the chromosome for spindle microtubule attachment. Instead, the whole chromosomal surface is bound and segregated. Genomic evidence suggests that many holocentric chromosomes lack tandem repeats and have cenH3 binding sites distributed over a wide variety of unique sequences throughout the chromosome, as expected. The nematode
Instead, the whole chromosomal surface is bound and segregated. Genomic evidence suggests that many holocentric chromosomes lack tandem repeats and have cenH3 binding sites distributed over a wide variety of unique sequences throughout the chromosome, as expected. The nematode
C. elegans
is the exemplar case of a holocentric organism. Recently, the chromosomal localization of histone cenH3 was determined in
C. elegans
by ChIP–chip analysis [93]. It was found that ~50% of the genome can be associated with cenH3, showing complete absence of particular DNA sequences to control cenH3 incorporation. Fundamental studies on the regulation of diffuse centromeres have been carried out on the nematode
Parascaris equorum
. In this organism, as in
C. elegans
, development is strictly mosaic, and each cell performs a predetermined number of cell divisions before differentiation. A consequence of this developmental system is that an induced death of a single cell can be lethal to the embryo because it cannot be replaced at the embryonic stage. Furthermore, in
Parascaris
, there is the phenomenon of chromatin diminution in somatic cells, which does not occur in germ cells. In embryonic somatic cells, both terminal and intercalary heterochromatin with no detectable kinetochore activity is eliminated by fragmentation, producing about 60 small chromosomes that are equipped with centromeres and that segregate correctly during mitosis [94]. In gonial cells, heterochromatin is not eliminated, and both heterochromatin and euchromatin retain kinetochore activity. Finally, in meiotic cells, centromeric activity is restricted to the terminal heterochromatic regions to which the spindle microtubules attach in the absence of a kinetochore plate. Therefore, in
Parascaris
, there is both structural and regional variability in relation to the cell type. In the embryo’s somatic cells, the centromeric activity is restricted to euchromatin, while in gonial cells it is diffused over different chromatin environments, and in meiotic cells is restricted to telomeric heterochromatin. Therefore,
Parascaris and other nematodes represent interesting models to study epigenetic centromere organization for the presence of three different centromere states under physiological conditions.
and other nematodes represent interesting models to study epigenetic centromere organization for the presence of three different centromere states under physiological conditions.
Conclusions
Proper segregation of chromosomes during cell division is essential for the survival of the cell and the whole organism. Therefore the centromere, a complex structure used for this function, has adapted during evolution to respond to changes in the cellular microenvironment as well to those in the external environment. The conflict between the need for functional stability and variability dictated by the environmental changes has been resolved with a wide flexibility through various epigenetic mechanisms. In order to keep the correct centromere functionality, epigenetic mechanisms can buffer possible centromere structural variations.
5. Conclusions
References
Proper segregation of chromosomes during cell division is essential for the survival of the cell and the whole organism. Therefore the centromere, a complex structure used for this function, has adapted during evolution to respond to changes in the cellular microenvironment as well to those in the external environment. The conflict between the need for functional stability and variability dictated by the environmental changes has been resolved with a wide flexibility through various epigenetic mechanisms. In order to keep the correct centromere functionality, epigenetic mechanisms can buffer possible centromere structural variations.
- Karpen, G.H.; Allshire, R.C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997, 13, 489-496.
- Earnshaw, W. C.; Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 1985, 91:313-321.
- Sullivan, K.F.; Hechenberger, M.; Masri, K. Human CENP-A Contains a Histone H3 Related Histone Fold Domain That Is Required for Targeting to the Centromere. Cell. Biol. 1994, 127, 581-592.
- Buchwitz, B. J.; Ahmad, K.; Moore, L.L.; Roth, M.B.; Henikoff, S. A histone-H3-like protein in elegans. Nature 1999, 401, 547–548.
- Takahashi, K.; Chen, E.S.; Yanagida, M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 2000, 288, 2215–2219.
- Howman, E.V.; Fowler, K.J.; Newson, A.J.; Redward, S.; MacDonald, A.C.; Kalitsis, P.; Choo, K.H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A 2000, 97, 1148–1153.
- Oegema, K.; Desai, A.; Rybina, S.; Kirkham, M.; Hyman, A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 2001, 153:1209–1226.
- Blower, M.D.; Karpen, G.H. The Role of Drosophila CID in Kinetochore Formation, Cell-Cycle Progression and Heterochromatin Interactions. Cell. Biol. 2001, 3, 730-739.
- Régnier, V.; Vagnarelli, P.; Fukagawa, T.; Zerjal, T.; Burns, E.; Trouche, D.; Earnshaw, W. Brown, W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Cell. Biol. 2005, 25, 3967–3981.
- McKinley, L.M.; Cheeseman, I.M. The molecular basis for centromere identity and function. Rev. Mol. Cell. Biol. 2016, 17:16–29.
- De Rop, V.; Padeganeh, A.; Maddox, P.S. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma 2012, 121, 527–538.
- Chen, C.C.; Mellone, B.G. Chromatin assembly: Journey to the CENter of the chromosome. Cell Biol. 2016, 214, 13–24.
- Jansen, L.E.T.; Black, B.E.; Foltz, D.R.; Cleveland, D.W. Propagation of centromeric chromatin requires exit from mitosis. Cell Biol. 2007, 176:795–805.
- Ahmad, K.; Henikoff, S. Centromeres are specialized replication domains in heterochromatin. J Cell. Biol. 2001, 153:101–110.
- Schuh, M.; Lehner, C.F.; Heidmann, S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Biol. 2007, 17, 237–243.
- Mellone, B.G.; Grive, K.J.; Shteyn, V.; Bowers, S.R.; Oderberg, I.; Karpen, G.H. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011, 7, e1002068.
- Dunleavy, E.M.; Beier, N.L.; Gorgescu, W.; Tang, J.; Costes, S.V.; Karpen, G.H. The Cell Cycle Timing of Centromeric Chromatin Assembly in Drosophila Meiosis Is Distinct from Mitosis Yet Requires CAL1 and CENP-C. PLoS Biol. 2012 10, e1001460.
- Lidsky, P.V.; Sprenger, F.; Lehner, C.F.; Distinct modes of centromere protein dynamics during cell cycle progression in Drosophila S2R+ cells. Cell Sci 2013, 126,4782-4793.
- Takayama, Y.; Sato, H.; Saitoh, S.; Ogiyama, Y.; Masuda, F.; Takahashi, K. Biphasic Incorporation of Centromeric Histone CENP-A in Fission Yeast. Biol. Cell. 2008, 19, 682–690.
- Cheng, Z.; Dong, F.; Langdon, T.; Ouyang, S.; Buell, C.R.; Gu. M.; Blattner, F.R.; Jiang, J. Functional Rice Centromeres Are Marked by a Satellite Repeat and a Centromere-Specific Retrotransposon. Plant Cell 2002 14, 1691-1704.
- Chang, C.H.; Chavan, A.; Palladino, J.; Wei, X.; Martins, N.M.C.; Santinello, B.; Chen, C.C.; Erceg, J.; Beliveau, B.J.; Wu, C.T.; Larracuente, A.M.; Mellone, B.G. Islands of retroelements are major components of Drosophila PLoS Biol. 2019, 17, e3000241.
- Chueh, A.C.; Northrop, E.L.; Brettingham-Moore, K.H.; Choo, K.H.A.; Wong, L, H. LINE Retrotransposon RNA Is an Essential Structural and Functional Epigenetic Component of a Core Neocentromeric Chromatin. PLoS Genet. 2009, 5, e1000354.
- Liu, Y.; Su, H., Zhang, J.; Liu, Y.; Feng, C.; Han, F. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol. 2020, 18, e3000582.
- Brown, J.B.; O’Neill, R. J. Chromosomes, Conflict, and Epigenetics: Chromosomal Speciation Revisited. Rev. Genomics Hum. Genet. 2010, 11, 291-316.
- Hartley, G.; O’Neill, R.J. Centromere Repeats: Hidden Gems of the Genome. Genes, 2019, 10, 223.
- Black, E.M.; Giunta, S. Repetitive Fragile Sites: Centromere Satellite DNA as a Source of Genome Instability in Human Diseases. Genes 2018, 9, 615.
- Guerrero, A.A.; Gamero, M.C.; Trachana, V.; Fütterer, A.; Pacio-Bras, C.; Dìaz-Concha, N.P.; Cogidosa, J.C.; Martínez-A.C.; van Welya, K.H.M. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Natl. Acad. Sci. USA 2010, 107, 4159–4164.
- Zeman, M.K.; Cimprich, K.A. Causes and Consequences of Replication Stress. Nat Cell Biol. 2014, 16, 2-9.
- Garavis, M.; Mendez-Lago, M.; Gabelica, V.; Whitehead, S.L.; Gonzales, C.; Villasante, A. The structure of an endogenous Drosophila centromeres reveals the prevalence of tandemly repeated sequences able to form i-motif. Sci Rep 2015, 5, 13307.
- Aze, A.; Sannino, V.; Soffientini, P.; Bachi, A.; Costanzo, Vi. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Cell Biol. 2016, 18, 684–691.
- Barra, V.; Fachinetti, D. The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Commun. 2018, 9, 4340.
- Valgardsdottir, R.; Chiodi, I.; Giordano, M.; Rossi, A.; Bazzini, S.; Ghigna, C.; Riva, S.; Biamonti, G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2007, 36, 423–434.
- Sengupta, S.; Parihar, R.; Ganesh, S. Satellite III Non-Coding RNAs Show Distinct and Stress-Specific Patterns of Induction. Biophys. Res. Commun. 2009, 382, 102–107.
- Hernández-Saavedra, D.; Strakovsky, R.S.; Ostrosky-Wegman, P.; Pan, Y.-X. Epigenetic Regulation of Centromere Chromatin Stability by Dietary and Environmental Factors. Nutr. 2017, 8, 889–904.
- Herr, I.; Debatin, K.M. Cellular stress response and apoptosis in cancer therapy. Blood 2001, 98, 2603-2614.
- Giunta, S.; Funabiki, H. Integrity of the Human Centromere DNA Repeats Is Protected by CENP-A, CENP-C, and CENP-T. Proc Natl Acad Sci U S A 2017, 114, 1928-1933.
- Gaubatz, J.W.; Cutler, R.G. Mouse Satellite DNA Is Transcribed in Senescent Cardiac Muscle. J Biol. Chem. 1990, 265(29):17753-8.
- Bouzinba-Segard, H.; Guais, A.; Francastel, C. Accumulation of Small Murine Minor Satellite Transcripts Leads to Impaired Centromeric Architecture and Function. Proc Natl Acad Sci U S A, 2006, 103(23):8709-14.
- Pezer, Z.; Ugarkovic, D. Satellite DNA-associated siRNAs as Mediators of Heat Shock Response in Insects. RNA Biol. 2012, 9(5):587-95.
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner; N.; Scheid, O. M.Epigenetic Regulation of Repetitive Elements Is Attenuated by Prolonged Heat Stress in Arabidopsis. Plant Cell, 2010, 22(9):3118-29.
- Tittel-Elmer, M.; Bucher, E.; Broger, L.; Mathieu, O.; Paszkowski, J.; Vaillant, I. Stress-induced Activation of Heterochromatic Transcription. PLoS Genet. 2010, 6, e1001175.
- Vourc'h, C.; Biamonti, G. Transcription of Satellite DNAs in Mammals. Mol. Subcell. Biol. 2011, 51, 95-118.
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barruca, M. Transcription of Tandemly Repetitive DNA: Functional Roles. Chromosome Res. 2015, 23(3):463-77.
- Grady, D. L.; Ratliff, R. L.; Robinson, D. L.; McCanlies, E. C.; Meyne, J.; Moyzis, R. K. Highly conserved repetitive DNA sequences are present at human centromeres. Natl. Acad. Sci. USA 1992, 89, 1695-1699.
- Jolly, C.; Metz, A.; Govin, J.; Vigneron, M.; Turner, B.M.; Khochbin, S.; Vourc’h, C. Stress-induced transcription of satellite III repeats. Cell Biol. 2004, 164, 25–33.
- Rizzi, N.; Denegri, M.; Chiodi, I.; Corioni, M.; Valgardsdottir, R.; Cobianchi F.; Riva, S.; Biamonti, G. Transcriptional Activation of a Constitutive Heterochromatic Domain of the Human Genome in Response to Heat Shock. Biol. Cell. 2004, 15, 543–551.
- Choi, E. S.; Strålfors, A.; Catania, S.; Castillo, A.G.; Svensson, J.P.; Pidoux, A.L.; Ekwall, K.; Allshine, R.C. Factors That Promote H3 Chromatin Integrity During Transcription Prevent Promiscuous Deposition of CENP-ACnp1 in Fission Yeast. PLoS Genet. 2012, 8,
- Carone, D.M.; Zhang, C.; Hall, L.E.; Obergfell, C.; Carone, B.R.; O’Neill, M.J.; O’Neill, R. Hypermorphic expression of centromeric retroelement-encoded small RNAs impairs CENP-A loading. Chromosome Res. 2013, 21, 49–62.
- Chan, F.L.; Wong, L.H. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 2012, 40, 11178–11188.
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Cell. 2017, 42, 226–240.
- Rošić, S.; Köhler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. Cell. Biol. 2014, 207, 335–349.
- Bobkov, G.O.M.; Gilbert, N.; Heun, P. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. Cell Biol. 2018, 217, 1957–1972.
- Jiang, J.; Birchler, J.A.; Parrott, W.A.; Dawe, R.K. A molecular view of plant centromeres. Trends Plant Sci. 2003, 8, 570-575.
- Topp, C.N.; Zhong, C.X.; Dawe, R.K. Centromere-encoded RNAs are integral components of the maize kinetochore. Natl. Acad. Sci. U S A, 2004, 101, 15986–15991.
- Lermontova, I.; Schubert, V.; Fuchs, J.; Klatte, S.; Macas, J.; Schubert, I. Loading of Arabidopsis Centromeric Histone CENH3 Occurs Mainly during G2 and Requires the Presence of the Histone Fold Domain. Plat Cell, 2006, 18, 2443–2451.
- Ohkuni, K.; Kitagawa, K. Role of transcription at centromeres in budding yeast. Transcription 2012, 3, 193-197.
- Feng, Q.; Moran, J.V.; Kazazian Jr, H.H.; Boeke, J.D. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 1996, 87(5):905-16.
- Jurka, J. Sequence Patterns Indicate an Enzymatic Involvement in Integration of Mammalian Retroposons. Proc Natl Acad Sci U S A 1997, 94(5):1872-7.
- Pimpinelli, S.; Berloco, M.; Fanti, L.; Dimitri, P.; Bonaccorsi, S.; Marchetti, E.; Caizzi, R.; Caggese, C.; Gatti, M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Natl. Acad. Sci. USA 1995 92(9):3804-8.
- Sun, X.; Le, H.D., Wahlstrom, J.M.; Karpen, G.H. Sequence Analysis of a Functional Drosophila Centromere. Genome Res 2003, 13(2):182-94.
- Neumann, P.; Yan, H.; Jiang, J. The Centromeric Retrotransposons of Rice Are Transcribed and Differentially Processed by RNA Interference. Genetics, 2007, 176, 749-761.
- De Cecco, M.; Criscione, S.W.; Peterson, A.L.; Neretti, N.; Sedivy, J.M.; Kreiling, J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissue. Aging 2013, 5, 867-883.
- Smurova, K.; De Wulf, P. Centromere and Pericentromere Transcription: Roles and Regulation… In Sickness and in Health. Genet. 2018, 9, 674.
- Klein, S. J.; O’Neill, R. J. Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 2018, 26, 5–23.
- Sullivan, B.A.; Karpen, G. H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Struct. Mol. Biol. 2004, 11, 1076–1083.
- Bergmann, J.H.; Rodríguez, M.G.; Martins, N.M.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.; Earnshaw, W.C. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. Embo J. 2011, 30, 328–340.
- Piacentini, L.; Marchetti, M.; Bucciarelli, E.; Casale, A.M.; Cappucci, U.; Bonifazi, P.; Renda, F.; Fanti, L. A role of the Trx-G complex in Cid/CENP-A deposition at Drosophila melanogaster centromeres. Chromosoma, 2019, 128, 503-520.
- Ohzeki, J.; Bergmann, J.H.; Kouprina, N.; Noskov, V.N.; Nakano, M.; Kimura, H.; Earnshaw. W.C.; Larionov, V.; Masumoto, H. Breaking the HAC Barrier: Histone H3K9 acetyl/ methyl balance regulates CENP-A assembly. EMBO J. 2012, 31, 2391–2402.
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; Choo, K.H.A. Centromere RNA Is a Key Component for the Assembly of Nucleoproteins at the Nucleolus and Centromere. Genome Res., 2007, 17, 1146-1160.
- Quenet, D.; Dalal, Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. ELife 2014, 3, e03254.
- Catania, S.; Pidoux, A.L.; Allshire, R.C. Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin. PLoS Genet. 2015, 11, e1004986.
- Liu, H.; Qu, Q.; Warrington, R.; Rice, A.; Cheng, N.; Yu, H. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Cell., 2015, 59, 426-36.
- Blower, M. D. Centromeric transcription regulates Aurora-B localization and activation. Rep. 2016, 15, 1624–1633.
- Grenfell, A.W.; Heald, R.; Strzelecka, M. Mitotic Noncoding RNA Processing Promotes Kinetochore and Spindle Assembly in Xenopus. J Cell Biol. 2016, 18, 133-141.
- Voullaire, L.E.; Slater, H.R.; Petrovic. V.; Andy Choo, K.H. A Functional Marker Centromere with No Detectable Alpha-Satellite, Satellite Ill, or CENP-B Protein: Activation of a Latent Centromere? J. Hum. Genet. 1993, 52, 1153-1163.
- du Sart, D.; Cancilla, M.R.; Earle, E.; Mao, J.; Saffery, R.; Tainton, K.M.; Kalitsis, P.; Martyn, J.; Barry, A.E.; Choo, K.H.A. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Genet. 1997, 16, 144-153.
- Choo K.H.A. Centromere DNA Dynamics: Latent Centromeres and Neocentromere Formation. J. Hum. Genet. 1997, 61, 1225-1233.
- Ventura, M.; Mudge, J.M.; Palumbo, V.; Burn, S.; Blennow, E.; Pierluigi, M.; Giorda, R.; Zuffardi, O.; Archidiacono, N.; Jackson, M.S.; Rocchi, M. Neocentromeres in 15q24-26 Map to Duplicons Which Flanked an Ancestral Centromere in 15q25. Genome Res. 2003, 13, 2059–2068.
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67.
- Platero, J.S.; Ahmad, K.; Henikoff, S. A Distal Heterochromatic Block Displays Centromeric Activity When Detached from a Natural Centromere. Cell 1999, 4, 995-1004.
- Amor, D.J.; Choo, K.H.A. Neocentromeres: Role in Human Disease, Evolution, and Centromere Study. J. Hum. Genet. 2002, 71, 695–714.
- Palladino, J.; Chavan, A.; Sposato, A.; Mason, T.D.; Mellone, B.G. Targeted De Novo Centromere Formation in Drosophila Reveals Plasticity and Maintenance Potential of CENP-A Chromatin. Cell 2020, 52, 379-394.
- Agudo, M.; Losada, A.; Abad, J.P.; Pimpinelli, S.; Ripoll, P.; Villasante, A. Centromeres from telomeres? The centromeric region of the Y chromosome of Drosophila melanogaster contains a tandem array of telomeric HeT-A- and TART-related sequences. Nucleic Acids Research, 1999, 27, 3318–3324.
- Villasante, A.; Abad, J.P.; and Mendez-Lago, M. Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. Proc Natl Acad Sci U S A 2007, 104, 10542-10547.
- Zeitlin, S.G.; Baker, N.M.; Chapados, B.R.; Soutoglou, E.; Wang, J.Y.J.; Berns, M.W.; Cleveland, D.W. Double-strand DNA Breaks Recruit the Centromeric Histone CENP-A. Natl. Acad. Sci. U S A 2009, 106, 15762-15767.
- Bodor, D.L.; Rodríguez, M.G.; Moreno, N.; Jansen, N.M.L. Analysis of Protein Turnover by Quantitative SNAP-based Pulse-Chase Imaging. Protoc. Cell Biol. 2012, 8, Unit8.8.
- Shang, W.H.; Hori, T.; Martins, N.M.C.; Toyoda, A.; Misu, S.; Monma, N.; Hiratani, I.; Maeshima, K.; Ikeo, K.; Fujiyama, A.; Kimura, H.; Earnshaw, W.C.; Fukagawa, T. Chromosome Engineering Allows the Efficient Isolation of Vertebrate Neocentromeres. Cell 2013, 24, 635–648.
- Nechemia-Arbely, Y.; Miga, K.H.; Shoshani, O.; Aslanian, A.; McMahon, M.A.; Lee, A.Y.; Fachinetti, D.; Yates, J.R.; Ren, B.; Cleveland, D.W. DNA Replication Acts as an Error Correction Mechanism to Maintain Centromere Identity by Restricting CENP-A to Centromeres. Cell. Biol., 2019, 21, 743-754.
- Liu, Y.G.; Whittier, R.F. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragment from PI and Yac clones for chromosomal walking. Genomics 1995, 25, 674-681.
- Li, T.H.; Schmid, C.W. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene 2001, 276, 135-141.
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Ratsch, G.; Weigel, D.; Laubinger, S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. The Plant Journal 2009, 58, 1068-1082.
- Fanti, L.; Piacentini, L.; Cappucci, U.; Casale, A.M.; Pimpinelli, S. Canalization by Selection of de Novo Induced Mutations. Genetics 2017, 206, 1995-2006.
- Gassmann, R.; Rechtsteiner, A.; Yuen, K.W.; Muroyama, A.; Egelhofer, T.; Gaydos, L.; Barron, F.; Maddox, P.; Essex, A.; Monen, J.; Ercan, S.; Lieb, J.D.; Oegema, K.; Strome, S.; Desai, A. An inverse relationship to germline transcription defines centromeric chromatin in elegans. Nat. 2012, 484, 534–537.
- Goday, C.; Pimpinelli, S. Chromosome Organization and Heterochromatin Elimination in Parascaris. Science, 1984, 224, 411-413.
