You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Daochen Zhu.
Dye decoloring peroxidases (DyPs) were named after their high efficiency to decolorize and degrade a wide range of dyes. DyPs are a type of heme peroxidase and are quite different from known heme peroxidases in terms of amino acid sequences, protein structure, catalytic residues, and physical and chemical properties. DyPs oxidize polycyclic dyes and phenolic compounds.
- dye decoloring peroxidase
- polycyclic dyes
1. Introduction
The dye decoloring peroxidases ((DyP, EC 1.11.1.19) the systematic name is reactive-blue-5: hydrogen-peroxide oxidoreductase) belongs to the heme peroxidases family [1]. In 1999, for the first time, a DyP was identified and purified from the basidiomycete Bjerkandera adusta. Since then, over the past 20 years, research on DyPs has grown exponentially [2]. DyPs are bifunctional enzymes with both oxidative and hydrolytic activities. It can degrade some tenacious substrates, including dyes, β-carotene, aromatic compounds, and sulfides. Fascinatingly, they prefer anthraquinone dyes as a substrate and have a high enzymatic activity for various organic compounds [3]. However, these are due to DyPs’ unique sequence and protein nature that differs from other known peroxidases [4]. Therefore, it has great application prospects in papermaking, coatings, environmental protection, bioenergy industries, etc.
In recent years, researchers focused on microbial DyPs and intensely observed their degrative supremacy, such as the degradation of dyes and lignin. Previous studies have clearly defined and explained the enzymatic structure and properties of DyPs, but its catalytic pathways for the degradation of dyes and lignin are still unclear. Although, DyPs have tremendous industrial and environmental applications. Unfortunately, several obstacles have hindered its industrial application, such as low industrial enzyme activity, foreign enzyme pollution, a complicated purification process, long growth cycle, and high cost. However, recently there have been several attempts to overcome these shortcomings and in the advancement in technologies and understanding of the catalytic pathways.
2. Composition and Structural Characteristics of DyPs
2.1. Sources and Classification of DyPs
Kim et al. (1999) discovered the first type of DyPs from fungi and named it for its ability to degrade various types of dyes. They characterized the first dye decolorizing protein in the fungus Thanatephorus cucumeris Dec 1 and isolated the first enzyme of the DyP family (BAD DyP) from the fungus [5]. Subsequently, various DyPs were isolated and characterized from different bacterial strains of DyP (Supplementary Table S1).
Peroxidase is an essential type of oxidoreductase, which contains heme protein, and an iron porphyrin IX as a prosthetic group exists in the active site. It is customary to divide heme peroxidases into two superfamilies: animal and plant peroxidase superfamilies. According to the homology of the primary structure, plant peroxidases are divided into categories I, II, and III, including fungal peroxidase (category II) and bacterial peroxidase (category I). Category I peroxidases have pigment C peroxidase, ascorbate peroxidase, and bacterial peroxidase [6].
Category II peroxidases are mainly peroxidases secreted by fungi, such as lignin peroxidase (LiP; EC 1.11.1.14), manganese peroxidase (MnP; EC 1.11.1.13), horseradish Peroxidase (HRP; EC1.11.1.7), and versatile peroxidase (VP; EC1.11.1.16) [7]. Enzymes of this category are extracellular and heme with different molecular characteristics such as the sequence of amino acids, protein 3D structure, catalytic activity, and environmental conditions. Some peroxidases secreted by plants, such as horseradish peroxidase and barley grain peroxidase, belong to the category III peroxidase [8]. The DyP forms a sandwich-shaped β sheet between the α helices. Traditional peroxidases are mainly composed of α helices and do not contain β sheets [9]. Studies have found that the DyPs enzyme has a high redox potential and can be used at pH 2–3. Anthraquinone dyes have high activity, while the other peroxidases do not have such characteristics [10]. Moreover, DyPs lacks plant peroxidase’s typical amino acid sequence (R-X-X-F/W-H), R represents arginine, and H represents distal histidine. Due to these differences between DyPs and other peroxidases, the DyPs enzyme should be a new type of heme peroxidase, different to other peroxidases [3]. According to the difference of origin, PeroxiBase classification database DyP-type peroxidases superfamily divided into four Classes A to D (http://peroxibase.toulouse.inra.fr, accessed on 4 July 2021). Different DyPs enzymes of A, B, C, and D show very similar protein tertiary structures (Z score > 20), and amino acid sequences of different types of DyPs are quite different. For example, the amino acid homology between Class D and Classes A, B, and C are 7%, 7%, and 16%, respectively [4]. Using the MATRAS tool to obtain the sequence alignment based on the protein tertiary structure of DyP, a new classification scheme for the DyPs family was proposed. This classification contains three classes (Class I, Class P, and Class V), where Class C and Class D were classified into Class V, while Class A and Class B are classified into Class I and Class P [4]. This kind of tertiary structure analysis is adequate for further analysis because they reveal the fundamental structural similarity between different DyP-type peroxidases. Currently, scientists are more inclined to use the PeroxiBase classification procedures to classify DyPs. However, there are only two enzymes in the Class C family whose structure has been elucidated [11,12][11][12].
2.2. Characteristics of DyPs
There are 237 enzymes in the DyPs family entered in the RedoxiBase database and DyP Class D accounts for more than 1/2, reaching 138. The first DyP-type enzymes discovered belong to the D-type DyPs (Figure 1). The D-type DyPs discovered are all produced by fungi, such as IlDyP produced by Irpex lacteus [13], rPsaDyP produced by Pleurotus sapidus [14], and AauDyP2 produced by Auricularia auricula-judae [15]. D-type DyP, as an extracellular enzyme, can be separated and purified from the microbial culture broth. Interestingly, these microorganisms often play a role in the degradation of lignin.
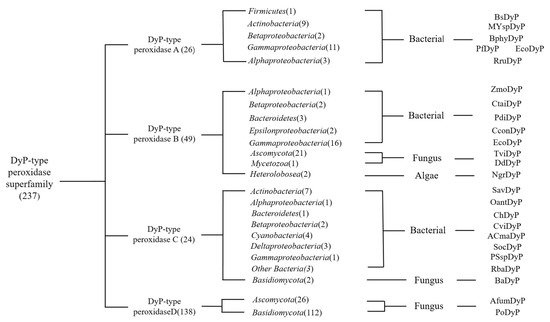

Figure 1. Phylogenic graph details the DyPs classes in the database and the corresponding producing microorganisms.
In contrast, Class A DyP was mainly isolated from bacteria. In Class B and C DyP, the primary source is still bacteria and lower eukaryotes produce a small part. It is worth noting that DyP from algae is found in Class B DyP. Among the microbes, the production cost of DyP from algae is lower than that of microbes when achieving higher yields. The length of the encoding amino acid sequence, encoding the D-type DyP, is longer, followed by the C, A, and B classes. Moreover, D-type DyP expresses a higher oxidation capacity in the catalytic efficiency of lignin model compounds and dyes, and the catalytic efficiency of AauDyP2 and Ildyp to Reactive Blue 5 and Reactive Black 5, which can reach 5.6 × 106 and 1.7 × 107 in (Kcat/Km) [13,15][13][15]. In addition, PsaPax also expresses high activity in olefin degradation [16]. Class C DyPs have various enzyme activities, such as DyP2 produced by actinobacteria Amycolatopsis sp. 75iv2. The electron transfer of hydrogen peroxide in the DyP2 catalytic process depends on the participation of Mn2+, and Mn2+ is regulated by the enzyme [11]. In the crystal structure of DyP2, it was observed that Mn2+ is combined and Mn2+ can greatly improve the catalytic efficiency.
On the other hand, Class B DyPs’ main characteristics are those specific protein-encoding genes and genes encoding capsular proteins are in operons form and form microcapsule structures that can protect DyPs [17]. In addition, some B-type DyPs, such as YfeX and YcdB, have the function of dechelating enzymes, which can extract an iron atom from heme without degrading the tetrapyrrole ring. The special features of Class A proteins are that some Class A proteins have two arginine ectopic (Tat) signal sequences at the N-terminus. It is confirmed that Class A enzymes rely on the TaT signal sequence to act outside the cytoplasm or the cell [18].
2.3. Structural Characteristics of DyP
Among the DyPs whose structures have been identified, including A, B, C, and D DyPs, it has been shown that it forms crystals in vitro with protoporphyrin (PPIX) at the heme-binding pocket except for EfeB [19]. DyPIn characterizes all DyPs and the binding pocket with an unclear function, and heme binds in the larger C-terminal region of the protein. In the wild-type DyP structure containing heme, the heme is coordinated by solvent molecules on the distal side, the coordination of type A DyPs occurs in the intermembrane zone, and the coordination of type C and D DyPs occurs at the C-terminal heme combination zone [20]. By observing the protein secondary structure of DyP, it was found that the heme progroup was located in the centre of the molecule, and a large number of α and β helix layers were folded. It is conceivable that the tertiary structure of DYP is mainly composed of dimeric or polymeric α-helix and β-helix. From the perspective of the tertiary structure of DyPs, the new classification method proposed by Yoshida and Sugano will significantly promote the study [4]. For example, according to the common structural characteristics of Class C and Class D in the same structural region, Class C DyP and Class D DyP can be classified as Class V. The important amino acid residues of Class P (Class B) DyPs are contained in the structural backbone. In contrast, the protein structures of Class A and Class V (Class C and Class D) have some extra regions based on Class P. This seems to explain, from another aspect, why Class C and Class D DyP can express higher catalytic efficiency because the function of enzymes often depends on its internal protein structure skeleton. Class C and Class D are larger than Class A and Class B from the structure size.
In addition, the binding residues of heme include conserved residues in DyPs and other residues in specific categories. In all DyPs enzymes, the proximal axial ligand of the heme iron structure is His, and it forms a hydrogen bond with the carboxylate of the acidic residue. There are also three residues at the distal end of heme iron: Asp, Arg, and Phe. These residues are similar in position to each other, while Arg is the closest to heme iron (NH1 to Fe—4.3 Å) and is compatible with propionic acid [21]. In addition, the polar residue (Asn246 in DypB) in the B-type DyPs hemoglobin pocket interacts with the iron-coordinated solvent. Although this residue is not conserved in other types of DyP, it corresponds to Ser331 in DyP2 and Gly348 in DyPdec1, and its function seems to be related to the distance between heme iron and the coordination solvent [22,23][22][23]. An interesting example is a novel cytoplasmic DyPA from Dictyostelium discoideum, which has the capability to oxidizes anthraquinone dyes and lignin model compounds. Unlike related enzymes, an aspartate residue replaces the first glycine of the conserved GXXDG motif in Dictyostelium DyPA. The active site of Dictyostelium DyPA has a hexa-coordinated heme iron with a histidine residue at the proximal axial position and either activated oxygen or a CN¯ molecule at the distal axial position [24].
2.4. The Physical and Chemical Properties of DyPs
Among all DyPs identified, except for a small number of bacteria that produce DyPs whose molecular weight is less than 40 kDa, the molecular weights of other enzymes are concentrated in 50–60 kDa, containing heme protein. There is an iron porphyrin IX at the active site [25]. DyPs mainly exist in the form of monomers, dimers, tetramers, and hexamers. Numerous studies have shown that DyP will maintain a high activity under acidic conditions, using 2,2-diaza-bis (3-ethyl-benzothiazole-6-sulfonic acid) diammonium salt (ABTS) as a substrate. In the enzyme activity test, the optimum pH of DyP is generally between 4.0 and 5.0. When SBP (soybean peroxidase) and CiP (generic peroxidases of Coprinopsis cinerea) are used as substrates, the optimum enzyme activity is 5.5 and 6.8 [15], respectively. For example, when BsDyP, PfDyP, and PpDyP are based on ABTS, 2,6-dimethoxyphenol, the optimal pH of DyP is mainly distributed between pH levels 2.0–4.0, 4.0–7.2, and 3.0–5.5, while the optimum pH varies [26]. In the practical application of DyPs, the optimal pH must ensure the high activity of DyP and consider the stability of DyP. Lier et al. have tested the pH catalytic effect on various DyPs and observed that although AauDyP1 has a high pH of 3.0 in an acidic environment, the activity is strong. At the same time, the stability of DyP is poor. Another study has observed that DyP has a stronger stability correlated with a lower activity when the pH value increases.
In addition, the temperature has a greater influence on enzyme activity. The optimal temperature of DyP is mainly concentrated at 40–60 °C [20]. Previously, the TfuDyP reaction rate was the highest at 45–60 °C at pH 5.5 utilizing DCP as a substrate, and the reaction rate decreases linearly as the temperature rises [27].
However, to use DyPs in industrial applications, it is crucial to coordinate the tolerance of pH and temperature and broaden the temperature and pH activity range. Various metal ions such as Ca2+, Mg2+, Zn2+, Fe2+, Mn2+, Co2+, Hg2+, Pb2+ and Pd2+ can also have an important impact on DyP activity. Previously, the effect of various ions on DyP B was investigated and it was found that Ca2+, Zn2+, and Mn2+ all make extracellular laccase activity and an increased effect of Zn2+ is the most significant [28]. Some metal ions such as Fe2+ and Hg2+ also have a strong inhibitory effect on DyP. Inhibition of Fe2+ may destroy the hydrogen peroxide transfer mechanism of DyP and make DyP itself active, while other ions may be in the process of reaction. These metal ions occupy the active centre of the DyP, change the structure of the DyP, and block the substrate’s binding with DyP, thereby inhibiting the enzyme activity. At the same time, Loncar also tested the effect of several inhibitors of aminotriazole, EDTA, imidazole, DTT, Cys, and sodium azide on enzyme activity. Under an inhibitory concentration of 0.5 mmol/L for DTT, Cys, and azide, the inhibitory effect of sodium sulfide on DyP was distinct, completely inhibiting the activity of DyP [20]. The inhibitory activity of aminotriazole is the lowest. DyP can still maintain around 60% of the activity. Still, the final complete inhibitory concentration of aminotriazole, EDTA, and imidazole has not been tested, and the tolerance of DyPs from different sources to inhibitors varies greatly and the tolerance of DyPs may also change accordingly after recombinant expression [27].
References
- Davies, M.J.; Hawkins, C.L.; Pattison, D.I.; Rees, M.D. Mammalian heme peroxidases: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 1199–1234.
- Heinfling, A.; Martínez, M.A.J.; Martínez, A.T.; Bergbauer, M.; Szewzyk, U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol. Lett. 1998, 165, 43–50.
- Sugano, Y.; Muramatsu, R.; Ichiyanagi, A.; Sato, T.; Shoda, M. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 2007, 282, 36652–36658.
- Yoshida, T.; Sugano, Y. A structural and functional perspective of DyP-type peroxidase family. Arch. Biochem. Biophys. 2015, 574, 49–55.
- Kim, S.J.; Shoda, M. Purification and characterization of a novel peroxidase from Geotrichum candidum dec 1 involved in decolorization of dyes. Appl. Environ. Microbiol. 1999, 65, 1029–1035.
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 871–897.
- Singh, S.; Pandey, V.P.; Naaz, H.; Dwivedi, U.N. Phylogenetic analysis, molecular modeling, substrate-inhibitor specificity, and active site comparison of bacterial, fungal, and plant heme peroxidases. Biotechnol. Appl. Biochem. 2012, 59, 283–294.
- Zámocký, M. Phylogenetic relationships in class I of the superfamily of bacterial, fungal, and plant peroxidases. Eur. J. Biochem. 2004, 271, 3297–3309.
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259.
- Yoshida, T.; Tsuge, H.; Konno, H.; Hisabori, T.; Sugano, Y. The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue. FEBS J. 2011, 278, 2387–2394.
- Brown, M.E.; Barros, T.; Chang, M.C. Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem. Biol. 2012, 7, 2074–2081.
- Ogola, H.J.; Kamiike, T.; Hashimoto, N.; Ashida, H.; Ishikawa, T.; Shibata, H.; Sawa, Y. Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 2009, 75, 7509–7518.
- Salvachúa, D.; Prieto, A.; Martínez, Á.T.; Martínez, M.J. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl. Environ. Microbiol. 2013, 79, 4316–4324.
- Lauber, C.; Schwarz, T.; Nguyen, Q.K.; Lorenz, P.; Lochnit, G.; Zorn, H. Identification, heterologous expression and characterization of a dye-decolorizing peroxidase of Pleurotus sapidus. AMB Express 2017, 7, 164.
- Liers, C.; Pecyna, M.J.; Kellner, H.; Worrich, A.; Zorn, H.; Steffen, K.T.; Hofrichter, M.; Ullrich, R. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl. Microbiol. Biotechnol. 2013, 97, 5839–5849.
- Krahe, N.K.; Berger, R.G.; Ersoy, F. A DyP-Type Peroxidase of Pleurotus sapidus with Alkene Cleaving Activity. Molecules 2020, 25, 1536.
- Contreras, H.; Joens, M.S.; McMath, L.M.; Le, V.P.; Tullius, M.V.; Kimmey, J.M.; Bionghi, N.; Horwitz, M.A.; Fitzpatrick, J.A.; Goulding, C.W. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J. Biol. Chem. 2014, 289, 18279–18289.
- Létoffé, S.; Heuck, G.; Delepelaire, P.; Lange, N.; Wandersman, C. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. USA 2009, 106, 11719–11724.
- Liu, X.; Du, Q.; Wang, Z.; Zhu, D.; Huang, Y.; Li, N.; Wei, T.; Xu, S.; Gu, L. Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes. J. Biol. Chem. 2011, 286, 14922–14931.
- Colpa, D.I.; Fraaije, M.W.; van Bloois, E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014, 41, 1–7.
- Zámocký, M.; Hofbauer, S.; Schaffner, I.; Gasselhuber, B.; Nicolussi, A.; Soudi, M.; Pirker, K.F.; Furtmüller, P.G.; Obinger, C. Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 2015, 574, 108–119.
- Zubieta, C.; Krishna, S.S.; Kapoor, M.; Kozbial, P.; McMullan, D.; Axelrod, H.L.; Miller, M.D.; Abdubek, P.; Ambing, E.; Astakhova, T.; et al. Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif. Proteins 2007, 69, 223–233.
- Singh, R.; Eltis, L.D. The multihued palette of dye-decolorizing peroxidases. Arch. Biochem. Biophys. 2015, 574, 56–65.
- Rai, A.; Klare, J.P.; Reinke, P.Y.; Englmaier, F.; Fohrer, J.; Fedorov, R.; Taft, M.H.; Chizhov, I.; Curth, U.; Plettenburg, O. Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum. Int. J. Mol. Sci. 2021, 22, 6265.
- Chen, C.; Shrestha, R.; Jia, K.; Gao, P.F.; Geisbrecht, B.V.; Bossmann, S.H.; Shi, J.; Li, P. Characterization of Dye-decolorizing Peroxidase (DyP) from Thermomonospora curvata Reveals Unique Catalytic Properties of A-type DyPs. J. Biol. Chem. 2015, 290, 23447–23463.
- Santos, A.; Mendes, S.; Brissos, V.; Martins, L.O. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: Towards biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2053–2065.
- Rahmanpour, R.; Rea, D.; Jamshidi, S.; Fülöp, V.; Bugg, T.D. Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch. Biochem. Biophys. 2016, 594, 54–60.
- Azevedo, A.M.; Martins, V.C.; Prazeres, D.M.; Vojinović, V.; Cabral, J.M.; Fonseca, L.P. Horseradish peroxidase: A valuable tool in biotechnology. Biotechnol. Annu. Rev. 2003, 9, 199–247.
More
